Identification and Genetic Characterization of a Novel Tillering Dwarf Semi-Sterile Mutant tdr1 in R
Vol.09No.13(2018), Article ID:88974,10 pages
10.4236/ajps.2018.913184
Bingrui Sun1,2*, Tingyou Huang3*, Chongyun Fu1,2#
1Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
2Guangdong Provincial Key Laboratory of New Technology in Rice Breeding, Guangzhou, China
3Mianyang Academy of Agricultural Sciences, Mianyang, China

Copyright © 2018 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: November 6, 2018; Accepted: December 2, 2018; Published: December 5, 2018
ABSTRACT
Tillering and plant height are important components of plant architecture and grain production in rice. We identified a novel high tillering, dwarf and semi-sterile mutant, as named tdr1 in a rice maintainer line E20 derived from the cross between between IR68888B and Luxiang 90. The investigation of tiller dynamic in the tdr1 line displayed 3 different phases: rapid increasing of tillers in the vegetative growth stage, producing no new tillers in the transition stage from the vegetative growth to reproductive growth, and regeneration of new tillers after heading. The assay of hormones showed the significant reduction of brassinolide level and no change of the levels of gibberellic acid, cytokinin and strigolactone in the tdr1 line. Genetic analysis indicated the phenotype of high tillering, dwarfism and semi-sterility is controlled by a recessive gene in several different segregation populations. The TDR1 gene was mapped in the 105 kb interval between RM3288 and RM6590 on chromosome 4. Cloning of TDR1 gene would provide a new opportunity to uncover the molecular mechanism of the development of plant height and tiller in rice.
Keywords:
Rice, High Tillering, Dwarf, Semi-Sterile, Mapping

1. Introduction
Plant height and tillering are two important traits of plant architecture affecting grain yields in rice. Dwarfism improves plant lodging resistance to increase grain yield and harvest index [1] . Rice tillering determines the number of panicles per plant. Excessive tillering generally causes a high rate of unproductive tiller, small panicle size and poor setting rate [2] . It was proved that there was a negative correlation between tiller number and plant height [3] .
Tillers originated from axillary buds in the axil of leaves and axillary buds are usually dormant after their formation. The outgrowth of axillary buds is regulated by the interaction of environmental and endogenous factors [4] . In rice, it is reported that the growth and development of tillers were associated with plant hormones such as auxin, cytokinin, gibberellins (GA), brassinosteroid and strigolactone [5] - [11] . A series of high-tillering dwarf mutants have been identified and characterized in rice in detail and some underlying genes have been cloned [12] - [17] .
In this study, we identified a novel mutant plant tdr1 with high-tillering, dwarf and semi-sterile phenotypes from a rice maintainer line E20 derived from the cross between IR68888B and Luxiang 90. We investigated its phenotype and the response to plant hormones and performed mapping analysis.
2. Materials and Method
2.1. Plant Materials
In the spring of 2006, we found a mutant with multi-tillers, dwarfism and semi-sterility (named as tdr) in the F4 progeny line E20 from the cross between IR68888B (female parent) and Luxiang 90 (male parent). We also created several F1populations derived from the crosses between the tdr1 line (female parent) and other indica lines E20, 931, IR68888B and Luxiang 90 (male parents).
2.2. Phenotypic Characterization and Assays of Phytohormone Level
In order to character the mutant phenotype, the mutant tdr, its wild type, and 9311 were grown in paddy field in Mianyang, Sichuan. A total of 30 plants from each of the above three lines were used to investigate several traits including tiller number, plant height, inter-node length, kilo-grain weight and pollen fertility.
Because the phenotypes of multi-tiller, dwarfism and semi-sterility were generally associated with plant hormones, exogenous gibberellins (1 × 10−4 mol/L GA3) and sterile water as control were sprayed for the tdr line, its wild type and 9311 at elongation stage in the greenhouse. The length of panicle, 1st, 2nd, 3rd, 4th and 5th upper internodes were measured for 10 plants each line at mature stage. Simultaneously, phytohormone levels were scored for young seedling using the ELISA kits (AndyGene) of four plant hormones including gibberellic acid (GA), cytokinin (CTK), brassinolide (BR) and strigolactone (SL) according to the corresponding protocols (http://www.andygene.com/index.php).
2.3. Extraction of DNA and Mapping of TDR Gene
DNA was prepared using the modified hexadecyltrimethylammonium (CTAB) method. Ten normal plants and 10 dwarf plants from the F2 population derived from tdr and 9311 were used to create the dominant and recessive bulks, respectively. Fine mapping of TDR gene was conducted with SSR and InDel markers. InDel markers were designed according to the genomic sequences of indica line 9311 and japonica line Nipponbare.
3. Results
3.1. Phenotypic Characterization of tdr1 Line
The mutant tdr showed dwarfism, high tillering ability, small grain and low setting rate (Figure 1). Ten plants from each lines tdr, its wild type and 9311 were characterized. Compared with the wild type, the maximum number of tillers in tdr1 line is up to 81.4 and its plant height is not enough half (45.5 cm) of the plant height of its wild type (109.86 cm). Its kilo-grain weight is 15.36 gram, the setting rate is only 32.47% and lots of unstained pollens were observed.
To fully understand tillering dynamics of tdr1 line, we continuously investigated the number of tillers of 10 plants for tdr1 line, its wild type and 9311 every 5 days after transplantation. We found that the tdr1 line has rapidly increased the tiller number since June 13th (Figure 2) and the number of tillers reached a plateau for the tdr1 line on July 8rd. For the wild type and 9311, the number of tillers have become immobile since June 28th, but new tillers occurred again on about August 7th (the heading day of tdr1 line) for the tdr1 line, which indicates that the transition from vegetative to reproductive stages could inhibit the growth of axillary buds and the end of the transition phase could relieve the dormancy of axillary buds in the tdr1 line again.
3.2. Effect Evaluation of Exogenous Gibberellin and Phytohormones Determination
In previous reports, GA plays an important role for plant height and tillering. In order to understand the effect of GA for the tdr1 line, we treated the tdr1 line, its wild type and 9311 with exogenous GA3 (1 × 10−4 mol/L). We found that exogenous GA can increase the plant height for the above three lines, but showed different effect for the three lines (Figure 3). For the tdr1 line, the 2nd and 3rd upper internodes were significantly elongated, but its plant height was not fully recovered. For its wild type, the length of the 1st, 2nd and 3rd upper internodes were remarkably increased. For 9311, the panicle, 1st, 2nd, 3rd and 4th internodes were significantly elongated. These above results indicate that the phenotype of the tdr1 line is not caused by the signal transduction pathway of GA.
To fully understand the change of phytohormones in the tdr1 line, we measured the content of phytohormones such as gibberellins (GA), cytokinin (CTK), brassinolide (BR) and strigolactone (SL) in the tdr line and its wild type. Compared with in the wild type (GA 113.02 pmol/L, CTK 41.99 pmol/L, SL 37.08 pmol/L), the levels of GA (109.34 pmol/L), CTK (43.05 pmol/L) and SL (35.18 pmol/L) did not significantly change in the tdr1 line, but the level of BR (165.22

Figure 1. The variant phenotype of the tdr1 line including plant height, the size of anthers and seeds and the fertility of pollen.

Figure 2. The dynamics of tiller number of the tdr1 line, its wild type and 9311.
pmol/L) remarkably decreased in the tdr1 line (Figure 4), implying the biosynthesis of brassinolide might play roles for the phenotype of tdr1 line.
3.3. Genetic Segregation of tdr Phenotype
To determine the inheritance pattern of the phenotype of high-tillering, dwarfism and semi-sterility in the mutant tdr1 line, we used the tdr1 line and four normal semi-dwarf lines with different genetic background such as its wild type E20, 9311, IR68888B and Luxiang 90 to construct several F2 populations and 1

Figure 3. Effects of exogenous GA on the length of different internodes and panicles in tdr1 line, its wild type and 9311.

Figure 4. The assays of four phytohormones including GAs, CTKs, BRs and SLs in the leaves of the tdr1 line and its wild type.
backcross population (BC1F1). We scored plant height of the parent lines, their F1, F2 and BC1F1 progenies and investigated the segregation ratios of plant height in F2 and BC1F1 populations (Table 1). All the F1 plants from the four crosses showed the wild-type phenotype, and all of these F2 progenies have a segregation ratio of 3:1 between wild-type and mutant plants (χ2 < 3.84), indicating this phenotype is controlled by a recessive gene.
3.4. The Mapping of TDR1 Gene
Genetic analysis showed that the phenotype of high-tillering, dwarfism and semi-sterility was controlled by a recessive gene. We selected the F2 population
Table 1. The phenotype of plant height in the different lines and progenies.
from the cross between 9311 and the tdr1 line to map the TDR gene. Thirty nine SSR markers showed the polymorphism between 9311 and tdr1 line in about 600 used SSR markers evenly distributed on 12 chromosomes in rice. We used these polymorphic SSR markers to perform the linkage analysis for 392 F2 recessive plants and finally the underlying gene TDR1 was located in the 2.1 cM interval between RM252 and RM303 on Chr.4 (Figure 5, Table 2).
For fine mapping of the TDR1 gene, we further screened the polymorphic markers between RM252 and RM303 based on the Nipponbare reference genome and obtained 7 polymorphic markers in this interval. The 1620 recessive plants in the F2 population were used to fine map the TDR1 gene. Finally, the TDR1 gene was mapped in the 105.4 kb interval between RM3288 and RM6590. Based on the Nipponbare reference genome (https://rapdb.dna.affrc.go.jp/), there are 20 annotated genes including D17/HTD1 (Os04g0550600) and OsSPL7 (Os04g0551500) (Table 3).
4. Discussion
Recently lots of high-tillering dwarf rice mutants were reported and some underlying genes were mapped and cloned. Phytohormones play important roles for plant growth and development. In this study, the tdr1 mutant displayed the capability of high tillering, dwarfism, small seeds and the reduction of fertility. The tdr1 line was treated with exogenous GA3 and its height was not fully recovered, which indicates the tdr1 mutant is independent of the GA pathway. The
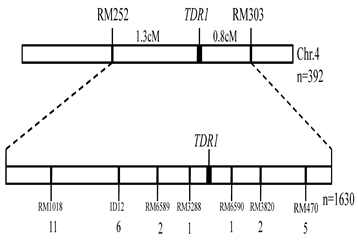
Figure 5. The linkage map of TDR1 gene.
Table 2. Lists of the primers used for mapping the underlying gene TDR1.
assay of phytohormone level also showed that this mutant phenotype was not related with GA, CTK and SL and could be caused by the reduction of BR. It is proved that BR plays crucial roles in the development of lateral organs including lateral organogenesis, plant height, seed size and fertility [18] [19] [20] . On the aspect of tiller dynamics, the tdr1 line rapidly increases of tillers in the vegetative growth phase, stops producing new tillers in the transition phase from the vegetative growth to reproductive growth, and regenerated new tillers after heading. The htd1 mutant has been reported to still keep the high tillering capacity [21] .
In our paper, the underlying gene TDR1 was located in the about 105 kb region of chromosome 4. In this candidate region, 20 genes are annotated and includes D17/HTD1 and OsSPL7 causing the phenotype of high tillering and dwarfism. D17/HTD1 was proved to be associated with strigolactone biosynthesis [22] [23] . However, our hormone assays also indicates that there is no significant change for strigolactone in the tdr1 line and its wild type. OsSPL7 was reported as a target of miR156f and binds directly the OsGH3.8 promoter to regulate tiller and plant height, and the miR156f/OsSPL7 pathway was involved in the regulation of plant architecture mediated by auxin [24] . Currently, it is
Table 3. The candidate genes in the mapping interval of TDR1 gene.
found that there is the interaction between auxin and brassinosteroid regulating plant growth and development. Auxin could promote the expression of DWARF4, a crucial hydroxylase for BR biosynthesis to control endogenous BR level and also inhibit the binding of BZR1 to the promoter of DWARF4 [25] [26] . In our study, the level of BRs is significantly reduced in the tdr1 line, compared with its wild type. Because the level of auxin was not tested, we can not know whether the auxin-BR interaction was destroyed to cause the phenotype of high tillering, dwarfism and semi-sterility in the tdr1 line.
Acknowledgements
This work was supported in part by grants from Sichuan Applied Basic Research Project (2015JY0061), Natural Science Foundation of Guangdong Province, China (2017A030310094) and Science and Technology Program of Guangzhou, China (201707010218).
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this paper.
Cite this paper
Sun, B.R., Huang, T.Y. and Fu, C.Y. (2018) Identification and Genetic Characterization of a Novel Tillering Dwarf Semi-Sterile Mutant tdr1 in Rice. American Journal of Plant Sciences, 9, 2545-2554. https://doi.org/10.4236/ajps.2018.913184
References
- 1. Marri, P.R., Sarla, N., Reddy, L.V. and Siddiq, E.A. (2005) Identification and Mapping of Yield and Yield Related QTLs from an Indian Accession of Oryza rufipogon. BMC Genetics, 6, 33-39. https://doi.org/10.1186/1471-2156-6-33
- 2. Peng, S., Khush, G.S. and Cassman, K.G. (1994) Evolution of the New Plant Ideotype for Increased Yield Potential. Workshop on Rice Yield Potential in Favorable Environments. International Rice Research Institute, Los Baños, 5-20.
- 3. Yan, J., Zhu, J., He, C., Benmoussa, M. and Wu, P. (1998) Quantitative Trait Loci Analysis for the Developmental Behavior of Tiller Number in Rice (Oryza sativa L). Theoretical and Applied Genetics, 97, 267-274. https://doi.org/10.1007/s001220050895
- 4. McSteen, P. and Leyser, O. (2005) Shoot Branching. Annual Review of Plant Biology, 56, 353-374. https://doi.org/10.1146/annurev.arplant.56.032604.144122
- 5. Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H. and Matsuoka, M. (2000) Rice Dwarf Mutant d1, Which Is Defective in the alpha Subunit of the Hetero-Trimeric G Protein, Affects Gibberellin Signal Transduction. Proceedings of the National Academy of Sciences, 97, 11638-11643. https://doi.org/10.1073/pnas.97.21.11638
- 6. Yamamuro, C., Ihara,Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H. and Matsuoka, M. (2000) Loss of Function of a Rice Brassinosteroid Insensitive1 Homolog Prevents Internode Elongation and Bending of the Lamina Joint. Plant Cell, 12, 1591-1606. https://doi.org/10.1105/tpc.12.9.1591
- 7. Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., Okabe, K., Kajiwara, H., Satoh, K., Yamamoto, K., Hirochika, H. and Kikuchi, S. (2002) Isolation and Characterization of a Rice Dwarf Mutant with a Defect in Brassinosteroid Biosynthesis. Plant Physiology, 130, 1152-1161. https://doi.org/10.1104/pp.007179
- 8. Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T., Kitano, H., et al. (2004). A Rice Semi-Dwarf Gene, Tan-Ginbozu (d35), Encodes the Gibberellin Biosynthesis Enzyme, Ent-Kaurene Oxidase. Plant Molecular Biology, 54, 533-547. https://doi.org/10.1023/B:PLAN.0000038261.21060.47
- 9. Tong, H. and Chu, C. (2012) Brassinosteroid Signaling and Application in Rice. Journal of Genetics and Genomics, 39, 3-9. https://doi.org/10.1016/j.jgg.2011.12.001
- 10. Jamil, M., Charnikhova, T., Houshyani, B., van Ast, A. and Bouwmeester, H.J. (2011) Genetic Variation in Strigolactone Production and Tillering in Rice and Its Effect on Striga hermonthica Infection. Planta, 235, 473-484. https://doi.org/10.1007/s00425-011-1520-y
- 11. Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., Yoshida, S., Ashikari, M., Kitano, H. and Matsuoka, M. (2003) A Rice Brassinosteroid-Deficient Mutant, Ebisu Dwarf (d2), Is Caused by a Loss of Function of a New Member of Cytochrome P450. Plant Cell, 15, 2900-2910. https://doi.org/10.1105/tpc.014712
- 12. Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I. and Kyozuka, J. (2005) Suppression of Tiller Bud Activity in Tillering Dwarf Mutants of Rice. Plant Cell Physiology, 46, 79-86. https://doi.org/10.1093/pcp/pci022
- 13. Zou, J.H., Zhang S.Y., Zhang, W.P., Li, G., Chen, Z.X., Zhai, W.X., Zhao, X.F., Pan, X.B., Xie, Q. and Zhu, L.H. (2006) The Rice HIGH-TILLERING DWARF1 Encoding an Ortholog of Arabidopsis MAX3 Is Required for Negative Regulation of the Outgrowth of Axillary Buds. The Plant Journal, 48, 687-696. https://doi.org/10.1111/j.1365-313X.2006.02916.x
- 14. Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., Sakakibara, H. and Kyozuka, J. (2007) DWARF10, an RMS1/MAX4/DAD1 Ortholog, Controls Lateral Bud Outgrowth in Rice. The Plant Journal, 51, 1019-1029. https://doi.org/10.1111/j.1365-313X.2007.03210.x
- 15. Gao, Z., Qian, Q., Liu, X., Yan, M., Feng, Q., Dong, G., Liu, J. and Han, B. (2009) Dwarf 88, a Novel Putative Esterase Gene Affecting Architecture of Rice Plant. Plant Molecular Biology, 71, 265-276. https://doi.org/10.1007/s11103-009-9522-x
- 16. Lin, H., Wang, R., Qian, Q., Yan, M., Meng, X., Fu, Z., Yan, C., Jiang, B., Su, Z., Li, J. and Wang, Y. (2009) DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. The Plant Cell, 21, 1512-1525. https://doi.org/10.1105/tpc.109.065987
- 17. Yao, R.F., Wang, L., Li, Y.W., Chen, L., Li, S.H., Du, X.X., Wang, B., Yan, J.B., Li, J.Y. and Xie, D.X. (2018) Rice DWARF14 Acts as an Unconventional Hormone Receptor for Strigolactone. Journal of Experimental Botany, 69, 2355-2365. https://doi.org/10.1093/jxb/ery014
- 18. Ryu, H. and Hwang, I. (2013) Brassinosteroids in Plant Developmental Signaling Networks. Journal of Plant Biology, 56, 267-273. https://doi.org/10.1007/s12374-013-0298-9
- 19. Zhang, C., Bai, M.Y. and Chong, K. (2014) Brassinosteroid-Mediated Regulation of Agronomic Traits in Rice. Plant Cell Reports, 33, 683-696. https://doi.org/10.1007/s00299-014-1578-7
- 20. Liu, F., Wang, P.D., Zhang, X.B., Li, X.F., Yan, X.H., Fu, D.H. and Wu, G. (2018) The Genetic and Molecular Basis of Crop Height Based on a Rice Model. Planta, 247, 1-26. https://doi.org/10.1007/s00425-017-2798-1
- 21. Zou, J.H., Chen, Z.X., Zhang, S.Y., Zhang, W.P., Jiang, G.H., Zhao, X.F., Zhai, W.X., Pan, X.B. and Zhu, L.H. (2005) Characterizations and Fine Mapping of a Mutant Gene for High Tillering and Dwarf in Rice (Oryza sativa L.). Planta, 222, 604-612. https://doi.org/10.1007/s00425-005-0007-0
- 22. Kulkarni, K.P., Vishwakarma, C., Sahoo, S.P., Lima, J.M., Nath, M., Dokku, P., Gacche, R.N., Mohapatra, T., Robin, S., Sarla, N., Seshashayee, M., Singh, A.K., Singh, K., Singh, N.K. and Sharma, R.P. (2014) A Substitution Mutation in OsCCD7 Cosegregates with Dwarf and Increased Tillering Phenotype in Rice. Journal of Genetics, 93, 389-401. https://doi.org/10.1007/s12041-014-0389-5
- 23. Yang, X., Chen, L., He, J. and Yu, W. (2017) Knocking out of Carotenoid Catabolic Genes in Rice Fails to Boost Carotenoid Accumulation, But Reveals a Mutation in Strigolactone Biosynthesis. Plant Cell Reports, 36, 1533-1545. https://doi.org/10.1007/s00299-017-2172-6
- 24. Dai, Z.Y., Wang, J., Yang, X.F., Lu, H., Miao, X.X. and Shi, Z.Y. (2018) Modulation of Plant Architecture by the miR156f-OsSPL7-OsGH3.8 Pathway in Rice. Journal of Experimental Botany, 69, 5117-5130. https://doi.org/10.1093/jxb/ery273
- 25. Chung, Y., Maharjan, P.M., Lee, O., Fujioka, S., Jang, S., Kim, B., Takatsuto, S., Tsujimoto, M., Kim, H., Cho, S., Park, T., Cho, H., Hwang, I. and Choe, S. (2011) Auxin Stimulates DWARF4 Expression and Brassinosteroid Biosynthesis in Arabidopsis. The Plant Journal, 66, 564-578. https://doi.org/10.1111/j.1365-313X.2011.04513.x
- 26. Yoshimitsu, Y., Tanaka, K., Fukuda, W., Asami, T., Yoshida, S., Hayashi, K., et al. (2011) Transcription of DWARF4 Plays a Crucial Role in Auxinregulated Root Elongation in Addition to Brassinosteroid Homeostasis in Arabidopsis thaliana. PLoS ONE, 6, e23851. https://doi.org/10.1371/journal.pone.0023851
NOTES
*The co-first authors contributed equally to this work.
上一篇:The Seed Water Sorption Isothe 下一篇:Cloning and Characterization o
最新文章NEWS
- Quantitative Screening of Secretory Protein Genes in Candidatus Liberibacter Asiaticus
- The Seed Water Sorption Isotherm and Antioxidant-Defensive Mechanisms of Hordeum vulgare L. Primed S
- Molecular Diversity in Selected Banana Clones (Musa AAA “Cavendish”) Adapted to the Subtropical Envi
- Nutritional Composition of Staple Food Bananas of Three Cultivars in India
- Physicochemical Properties and Nutritional Ingredients of Kernel Oil of Carya cathayensis Sarg
- Ecofriendly Remediation of Pulp and Paper Industry Wastewater by Electrocoagulation and Its Applicat
- In Vitro Plant Regeneration of Dendrocalamus stocksii (Munro) M. Kumar, Remesh & Unnikrisnan, throug
- Evaluation for Innovation Ability of National Agricultural Science and Technology Parks in Jiangxi P
推荐期刊Tui Jian
- Chinese Journal of Integrative Medicine
- Journal of Genetics and Genomics
- Journal of Bionic Engineering
- Pedosphere
- Chinese Journal of Structural Chemistry
- Nuclear Science and Techniques
- 《传媒》
- 《中学生报》教研周刊
热点文章HOT
- Genetic Analysis of Selected Mutants of Cowpea (Vigna unguiculata [L.] Walp) Using Simple Sequence R
- DNA Barcoding and Identification of Medicinal Plants in the Kingdom of Bahrain
- Cloning and Characterization of a Mitogen-Activated Protein Kinase Gene 84KMPK14 in Hybrid Poplar (P
- Morphological Investigation of Genus Ziziphus Mill. (Rhamnaceae) in Saudi Arabia
- Effect of Plant Extracts on Seed Borne Fungi of Jute
- Molecular Diversity in Selected Banana Clones (Musa AAA “Cavendish”) Adapted to the Subtropical Envi
- Evaluation for Innovation Ability of National Agricultural Science and Technology Parks in Jiangxi P
- Correlation and Path Coefficient Analyses of Cowpea (Vigna unguiculata L.) Landraces in Ethiopia

