Quantitative Gene Expression of Peroxidase, Polyphenoloxidase and Catalase as Molecular Markers for
Vol.06No.02(2016), Article ID:65309,13 pages
10.4236/ajmb.2016.62010
Eman El-Argawy1, Ibrahim A. Adss2
1Plant Pathology Department, Faculty of Agriculture, Damanhour University, Damanhour, Egypt
2Division of Genetics, Faculty of Agriculture, Damanhour University, Damanhour, Egypt

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 19 January 2016; accepted 2 April 2016; published 5 April 2016
ABSTRACT
Brown rot or bacterial wilt of potato caused by Ralstonia solanacearum, is an economically important disease. Potato, cv. Nicola, was found to be relatively highly resistant to the infection with R. solanacearum and showed 15.12% wilt disease index, meantime, cv. Kara showed intermediate resistance with 37.40% disease index while, cv. Spunta was susceptible and showed 80.33% disease index. The role of defense-related enzymes in imparting resistance in potato against R. solanacearum was investigated by quantifying enzymes activity and gene expression of three defense- related enzymes, peroxidase, polyphenol oxidase and catalase. Peroxidase showed maximum activity 0.488 min−1∙g−1 early at 12 h after pathogen inoculation in the cv. Nicola, whereas in susceptible cultivar Spunta showed lower activity of maximum 0.226 min−1∙g−1 later at 48 h after inoculation. While, the moderately resistant cultivar Kara showed intermediate activity for the peak and its time. Meanwhile, polyphenol oxidase showed similar trends to that of peroxidase. On the contrary, catalase showed the highest activity values in the susceptible, cv. Spunta, while, in relatively highly resistant (cv. Nicola) and the moderately resistant (cv. Kara) showed lower values of activity and up to 96 h after inoculation. Meanwhile, gene expression of related enzymes the RT-PCR was used. At zero time, the relatively highly resistant potato cultivar, Nicola, showed the highest values of gene expression for both Peroxidase (POD) and Poly Phenol Oxidase (PPO). While, the susceptible potato cultivar, Spunta showed the lowest values. On the contrary, Catalase (CAT) gene expression was the highest in the susceptible, cv. Spunta, and was the lowest in the relatively highly resistant, cv. Nicola, while, was of intermediate values in the intermediate resistance, cv. Kara. Results show that peroxidase and polyphenol oxidase activities can be used as biochemical markers to reveal the resistance and susceptibility nature of potato cultivars against bacterial wilt disease of potato caused by R. solanacaerum.
Keywords:
RT-PCR, Defense Related Enzymes, Gene Expression, Potato Brown Rot, Ralstonia solanacearum

1. Introduction
Potato (Solanum tuberosum L.) is one of the most consumed crops in the world with global production of approximately 367,753,014 tons, produced from approximately 19,454,997 hectares [1] around the world. Meantime, it is considered one of the most important vegetable crops in Egypt. Potato production is approximately 4,800,000 tons, production from approximately 178,000 hectares, which making Egypt Africa’s No. 1 potato producer [1] . However, brown rot or bacterial wilt of potato caused by Ralstonia solanacearum constitutes a threat to potato cultivation in Egypt and in all tropical agriculture [2] . R. solanacearum has a large host range of more than 200 species in 50 families [3] and affects a wide range of economically important crops such as tomato, potato, eggplant, chili and non solanaceous crops such as banana and groundnut [4] . Bacterial wilt causes 15% to 55% crop losses around the world. Disease resistance in plants is associated with activation of a wide array of defense responses that slow down or halt infection at certain stages of the host-pathogen interaction. These defense mechanisms include preexisting physical and chemical barriers that interfere with pathogen establishment. In response to the infection, the host induces a cascade of pathogen inducible enzymes, which are implemented in defense against phytopathogens. Early and elevated levels of expressions of various defense enzymes are an important feature including peroxidase (POD), polyphenol oxidase (PPO) and catalase (CAT) during host pathogen interactions. Polyphenol oxidase (PPO) is one of the main SAR related enzymes in plant, and their activities are related to plant resistance [5] [6] . Increases in POD activity are often associated with a progressive incorporation of phenolic compounds within the cell wall during incompatible plant-microbe/elicitor interactions. CAT is an antioxidative enzyme involved in oxidative burst generated transiently in plant-pathogen interactions. CAT is involved in regulation of H2O2 levels in plant tissues [7] . Real Time PCR (RT-PCR) has become an extensively applied technique in molecular plant pathology and it has been extensively used for quantification of different enzymes gene expression activities [8] . RT-PCR proved to be a successful tool for the evaluation of possible resistance in wheat [9] .
Therefore aims of the present study were to:
1) Investigate the reaction of different potato cultivars to infection with R. solanacearum.
2) Investigate the potential of peroxidase (POD) and catalase (CAT) enzymes activity in relation to potato resistance to R. solanacearum.
3) Quantifying the gene expression levels of peroxidase, polyphenol oxidase and catalase enzymes using Real Time PCR.
2. Materials and Methods
2.1. Plant and Fungal Materials
Certified tubers of cvs.Spunta, Nicola and Kara potato were purchased from the International Potato Center (CIP) Kafr El-Zayat, Egypt. These three potato varieties were previously proved to have different degree of susceptibility against R. solanacearum.
A highly virulent isolate of R. solanacearum was obtained from R. solanacearum collection bank, Plant Pathology Dept., Faculty of Agriculture, Damanhour University.
Potato tubers of the three cultivars Spunta, Kara and Nicola were surface sterilized with 1% sodium hypochlorite for five minutes, washed with sterile water, and planted in plastic pots 30 cm diameter filled with sterile peat moss (one tuber per pot). When plant reached 15 - 20 cm tall (25 days after plantation), stems were inoculated by injecting by a sterilized needle 0.25 ml, bacterial suspension (109 cfu/ml) into the stems at 5 cm above the soil level according to (Prior and Steva, 1990) [10] . Ten replicates were used in the experiment and plants injected with sterile distilled water were served as control. Inoculated plants were placed in a greenhouse at 25˚C ± 2˚C.
2.2. Disease Assessments
After 5 weeks of inoculation, wilt severity was assessed according to [11] as follows: 0:extremely resistant, no wilting; 1: highly resistant, 1% - 25% of total leaves wilted; 2: moderately resistant, 26% - 50% of total leaves wilted; 3: susceptible, 51% - 75% of total leaves wilted and 4: highly susceptible, 76% - 100% of total leaves wilted or plant died. Disease severity as disease index was calculated according to the formula:

where ni = number of plants with respective disease rating; vi = disease rating; V = the highest disease rating; and N = the number of plants observed.
2.3. Assessment of Potato Growth Parameter
Five weeks after inoculation plants were uprooted and examined for the following growth parameters:
Shoot fresh weight (g).
Root fresh weight (g).
Total tubers weight/plant (g).
Then, percentage of reduction according to the control was calculated in each potato cultivar.
2.4. Determination of Photosynthetic Pigments
Fresh leaves samples of inoculated and non-inoculated potato cultivars were collected after 5 weeks from inoculation for estimation of photosynthetic pigments.
Chlorophyll A, B and β-carotene were determined according to [12] as follows: half gram fresh leaves were ground in a pestle and mortar and extracted by 15 ml of 80% acetone (1:100 w/v) and 0.5 g calcium carbonate. The mixture was filtered through a glass funnel and the residue was washed with a small volume of acetone and completed to 25 ml. The optical density (O.D) of a constant volume of filtrate was measured at a wave length of 662 nm, 644 nm and 440 nm for chlorophyll A, chlorophyll B and carotene, respectively. The experiment was repeated thrice.
The following equations were used:
Chl.A = 9.784 E.662 − 0.99 E.644 = mg/L
Chl.B = 21.426 E.644 − 4.65 E.662 = mg/L
Carotene = 4.695 E.440 − 0.268 (Chl.A − Chl.B) = mg/L
where, E. = Optical density at the wavelength indicated.
2.5. Determination of the Defense Related Enzyme Activity in Potato Cultivars against R. solanacearum
2.5.1. Estimation of Peroxidase Activity
The peroxidase (POD) activity was assayed as described by [13] . The peroxidase activity was measured at 0, 3, 6, 12, 24, 48, 72 and 96 hours (h) after inoculation. Extraction was carried out by homogenizing 1 g of the fresh leaves of inoculated potato samples in 2.6 mL of 0.1 M sodium phosphate buffer (pH 6.5) using pre chilled pestle and mortar (4˚C). The homogenate was centrifuged at 10,000 rpm for 15 mins at 4˚C. The supernatant was served as enzyme source for the reaction mixture which consisted of 1.5 mL of 0.05 M pyrogallol, 0.5 mL of enzyme extract, and 0.5 mL of 1% H2O2. The reaction mixture was incubated at 28˚C ± 2˚C. At the start of enzyme reaction, the absorbance of the mixture was set to zero at 420 nm in the spectrophotometer and the change in the absorbance was recorded at 20 s intervals for 3 mins. Boiled enzyme preparation was served as a control. Peroxidase activity was expressed as change in the absorbance of the reaction mixture min−1∙g−1 protein of fresh tissue. All the experiments were repeated thrice.
2.5.2. Estimation of Polyphenol Oxidase Activity
One gram of the leaf sample was homogenized in 2 mL of 0.1 M sodium phosphate buffer (pH 6.5) in a pre- chilled pestle and mortar. The homogenate was centrifuged at 10,000 rpm for 15 min at 4˚C and the supernatant served as enzyme source. Polyphenol oxidase activity was determined according to the procedure given by [14] . The reaction mixture consisted of 1.5 mL of 0.1 M sodium phosphate buffer (pH 6.5) and 200 µL of the enzyme extract. To start the reaction, 200 µL of 0.01 M catechol was added. The reaction mixture was incubated at room temperature and the absorbance was set to zero at 495 nm. The changes in absorbance were recorded at 30 s interval for 2 min and the activity was expressed as change in absorbance min−1∙g−1 of fresh tissue. The polyphenol oxidase activity was measured at 0, 3, 6, 12, 24, 48, 72 and 96 hours after inoculation. All the experiments were repeated thrice.
2.5.3. Estimation of Catalase (CAT) Activity
Catalase (CAT) activity was measured according the methods given by [15] with a slight modification. Extraction was carried out by homogenizing 1 g of the fresh leaves of inoculated potato samples in 2.6 mL of 0.1 M sodium phosphate buffer (pH 6.5) using pre chilled pestle and mortar (4˚C). The assay mixture contained 2.6 mL of 50 mM potassium phosphate buffer (pH 7.0), 0.4 mL of 15 mM H2O2 and 0.04 mL of the enzyme extract. The extract was centrifuged at 4˚C for 20 min at 12,500 rpm. The supernatant was used for enzyme assay. The decomposition of H2O2 was followed by the decline in absorbance at 240 nm. The enzyme activity was expressed in mixture min−1∙g−1 protein. The catalase activity was measured at 0, 3, 6, 12, 24, 48, 72 and 96 hours after inoculation. All the experiments were repeated thrice.
2.6. Quantification of the Enzyme Genes Expression Associated with Resistance to R. solanacearum
2.6.1. RNA Isolation and Preparation for RT-PCR
Total RNA was extracted from plant tissue using Green Start™ RNA Isolation kit ІІ (guanidiumthiocynate) according to the manfacture procedures.
Reverse transcription (RT) or first strand reaction was performed for converting the mRNA to complementary DNA (cDNA) in the presence of dNTPs (deoxynucleotide triphosphates) reverse transcriptase. The components are combined with a DNA primer in a reverse transcriptase buffer for an hour at 42˚C. The exponential amplification via reverse transcription polymerase chain reaction provides a highly sensitive technique, where a very low copy number of RNA molecules can be detected.
Reverse transcription reaction was performed using oligo (dT) primer (5’-TTTTTTTTTTTTTTT-3’). Each 25 µl reaction mixture contained 2.5 µl (5×) buffer with MgCl2, 2.5 µl (2.5 mM) dNTPs, 1 µl (10 pmol) primer, 2.5 µl RNA (2 mg/ml) and 0.5 unit reverse transcriptase enzyme. PCR amplification was performed in a thermal cycler programmed at 42˚C for 1 hr, 72˚C for 10 min (enzyme killing) and the product was stored at 4˚C until use.
2.6.2. Estimation of Quantitative Enzyme Gene Expression Using RT-qPCR
According to [16] with some modifications. Samples were analyzed using the Fermentasekit: Each reaction contained 12.5 μl of 2× Quanti tech SYBR® Green RT Mix, 1 μl of 25 pm/μl forward primer, 1 μl of 25 pm/μl reverse primer,1 μl of the cDNA (50 ng), 9.25 μl of RNase free water for a total of 25 μl. Samples were spun before loading in the Rotor’s wells.
The real time PCR program was as follows: initial denaturation at 95˚C for 10 min; 40 cycles of at ˚C for 15 sec.; annealing at 60˚C for 30 sec and extension at 72˚C for 30 sec. Data acquisition performed during the extension step. This reaction was performed using Rotor-Gene-6000-system (Qiagen, USA) with a set of primers for peroxidase, polyphenol oxidase and catalase genes (Table 1).
Table 1. Sequence of primers used in the real-time PCR.
(F) Forward primer; (R) Reverse primer.
2.6.3. Data Analysis
Comparative quantification analysis was done using Rotor-Gene-6000 Series Software according to [17] .
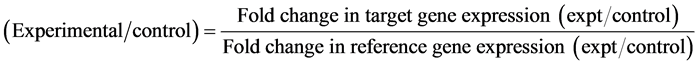
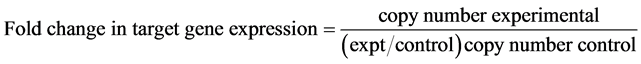

The equation shows a mathematical model of relative expression ratio in real-time PCR. The ratio of a target gene is expressed in a sample versus a control in comparison to a reference gene.
The sample and control dataset of real-time PCR data were analyzed with appropriate Bioinformatics and Statistical program for the estimation of the relative expression of genes using real-time PCR and the result normalized to ITS housekeeping gene (Reference gene). The data were statistically evaluated, interpreted and analyzed using Rotor-Gene-6000 version 1.7.
2.7. Statistical Analysis
The obtained data were statistically analyzed using the American SAS/STAT Software, version 6 and means were compared by the least significant difference test (LSD) [18] .
3. Results
3.1. Susceptibility of Different Potato Cultivars to R. solanacearum
Data in (Table 2) shows that potato, cv. Nicola was the most resistant potato cultivar to R. solanacearum and shows disease index as low as 15.12%. Meantime, cv. Kara, showed moderate resistance as exhibited 37.40% wilt disease index. On the other hand, cv. Spunta was highly susceptible and showed 80.33% disease index to the infection with R. solanacearum the incitant of the potato bacterial wilt disease (Table 2).
3.2. Potato Growth Parameters
Data in (Table 3), illustrated in (Figure 1) showed that there was no significant variations among the tested non inoculated (healthy) potato cultivars for the assessed vegetative traits, i.e. shoot fresh weight/plant, root fresh weight/plant and total tuber fresh weight/plant. However, after the inoculation with R. solanacearum, the tested potato cultivars showed significant variations for the tested vegetative traits. Potato, cv. Nicola, showed the most resistance expressed in terms of the lowest reduction in shoot fresh weight/plant (11.74%), root fresh weight/ plant (13.71%) and total tuber fresh weight/plant (12.59%). This comparing to high percentage of reductions in the most susceptible potato, cv. Spunta, being 63.98%, 71.25% and 62.68% for the same traits, respectively. Meanwhile, cv. Kara showed intermediate values for the previous traits being 30.54%, 43.29% and 40.54%, respectively.
Table 2. Wilt disease index (severity) developed on potato plants of different cultivars artificially inoculated with R. solanacearum under greenhouse condition, five weeks after inoculation.
Data are means of ten replicates.


Figure 1. Shoot fresh weight/plant, root fresh weight/plant and total tuber fresh weight/plant of three potato cultivars (Nicola, Spunta and Kara) inoculated with R. solanacearum, under greenhouse conditions, five weeks after inoculation.
Table 3. Values of vegetative traits of three potato cultivars artificially inoculated with R. solanacearum under greenhouse conditions five weeks after inoculation.
Data are means of four replicates; growth parameters were assessed five weeks after potato inoculation with R. solanacearum; values followed by different letters for each parameter are significantly different at p = 0.05.
3.3. Potato Chlorophyll Content
Data in (Table 4), illustrated in (Figure 2) shows that, there was no significant variations between healthy non- inoculated plants of cv. Nicola (relatively highly resistant) and cv. Spunta (susceptible) for the measured chlorophyll contents, chlorophyll A, chlorophyll B and the carotene. However, after the inoculation with R. solanacearum, considerable variations were revealed where the inoculationwith R. solanacearum resulted in significant decrease in all pigment contents, chlorophyll A, chlorophyll B and the carotene. Meantime, cv. Spunta exhibited the lowest pigment contents being 0.05, 0.11 and 0.1 for chlorophyll A, chlorophyll B and the carotene, respectively. This comparing to 0.09, 0.16 and 0.28 for the same pigments in cv. Nicola (relatively high resistance). While, for most pigments there was no significant differences between cv. Spunta (susceptible) and cv. Kara (intermediate resistance).


Figure 2. Effect of artificial inoculation with R. solanacearum on chlorophyll A, chlorophyll B and carotene contents in three potato cultivars tested, five weeks after inoculation under greenhouse conditions.
Table 4. Effect of artificial inoculation with R. solanacearum on chlorophyll A, chlorophyll B and carotene contents in the three tested potato cultivars, five weeks after inoculation under greenhouse conditions.
Data are means of three replicates. Photosynthetic pigments were examined five weeks after potato inoculation with R. solanacearum. Values followed by different letters for each parameter are significantly different at p = 0.05.
3.4. Potato enzyme Activity Associated with Resistance to R. solanacearum
Peroxidase (POD), polyphenol oxidase (PPO) and catalase (CAT) activities were assessed spectrophotometrically in the potato tuber tissues after inoculation with R. solanacearum. Data illustrated in (Figure 3) showed that, in the relatively highly resistant cultivar (Nicola) a drastic increase in peroxidase (POD) activity was noticed after inoculation and reached its peak (0.488 min−1∙g−1) soon at 12 h, and kept at the higher level up to 72 h after inoculation. On the contrary, in the susceptible, cv. Spunta, the POD activity peak was as low as 0.226 min−1∙g−1 and was recognized later at 48 h, after inoculation (Figure 3). Potato, cv. Kara, however, of the intermediate resistance to R. solanacearum, showed intermediate POD activity for the peak value and its time.
Concerning polyphenol oxidase (PPO) activity, in relatively highly resistant, cv. Nicola, the peak activity of PPO was the highest (0.445 min−1∙g−1) at 24 h, and kept at high level up to 72 h after inoculation. However, in susceptible, cv. Spunta, the maximum activity was 0.364 min−1g−1 and was recognized as later at 48 h after inoculation. Meanwhile, cv. Kara, of the intermediate resistance showed intermediate values (Figure 3).
On the contrary, catalase (CAT) activity, in the susceptible, cv. Spunta, was the highest and remained at the high levels up to 96 h after inoculation, while, in relatively highly resistant one, cv. Nicola, and moderately resistant one, cv. Kara, showed lower values of activity.
3.5. Quantitative Enzyme Gene Expression Associated with Potato Resistance to R. solanacearum
The relative expression of peroxidase (POD), polyphenol oxidase (PPO) and catalase (CAT) genes were quantified by real-time reverse transcription RT_qPCR analysis at 0, 3, 6, 12, 24, 48, 72 and 96 hours after inoculation of the three tested potato cultivars, Nicola, Kara and Spunta with R. solanacearum. Data illustrated in (Figure 4) shows that, at zero time, the relatively highly resistant potato cv. Nicola showed the highest values of gene expression for both POD and PPO. However, the susceptible potato cultivar, Spunta, showed the lowest values, while, the intermediate resistance potato cultivar, Kara, exhibited intermediate values.
Meanwhile, POD and PPO exhibited their peaks sooner at 12 h and 24 h, respectively, after inoculation of the relatively highly resistant potato cultivar, Nicola, while for the susceptible, cv. Spunta, the POD and PPO peaks were recognized later at 48 h and with considerable low level.
On the contrary, for CAT, gene expression was the highest in cv. Spunta (susceptible) and was the lowest in, cv. Nicola, (relatively highly resistant) while, it was of intermediate values in cv. Kara (intermediate resistance) (Figure 4).


Figure 3. Peroxidase (POD), polyphenol oxidase (PPO) and catalase (CAT) activity in potato cultivars, after inoculation with R. solanacearum.


Figure 4. Peroxidase (POD), polyphenol oxidase (PPO) and catalase (CAT) gene expression in potato cultivars, after inoculation with R. solanacearum.
4. Discussion
In the present study potato, cv. Nicola was the relatively highly resistant potato cultivar to R. solanacearum and showed disease index as low as 15.12%. Meantime, cv. Kara was shown to be moderately resistant as exhibited 37.40% wilt disease index; on the other hand, cv. Spunta, was susceptible and showed 80.33% disease index to the bacterial wilt disease. The study indicated that there was no significant variations among the tested uninoculated (healthy) potato cultivars for the assessed vegetative traits, shoot fresh weight/plant, root fresh weight/plant and total tuber fresh weight/plant. However, after the inoculation with R. solanacearum, the tested potato cultivars
showed significant variations for the assessed vegetative traits. Potato plants, cv. Nicola, showed the relatively highly tolerance expressed in terms of the lowest reduction in shoot fresh weight/plant (11.74%), root fresh weight/plant (13.71%) and total tuber fresh weight/plant (12.59%). This in comparison with high percentage of reductions in the susceptible potato, cv. Spunta, being 63.98%, 71.25% and 62.68% for the same traits, respectively. Meanwhile, cv. Kara, showed intermediate values for the previous traits being 30.54%, 43.29% and 40.54%, respectively.
There was no significant difference in the uninoculated healthy potato plants between, cv. Nicola, (relatively highly resistant) and, cv. Spunta, (susceptible) for the measured pigments contents, chlorophyll A, chlorophyll B and the carotene. However, after the inoculation with R. solanacearum, more variations were revealed where, cv. Spunta, exhibited the lowest pigment contents. While, cv. Nicola (relatively highly resistant) exhibited higher pigment contents. Meanwhile, for most pigments there was no significant difference between cv. Spunta (susceptible) and cv. Kara (intermediate resistance).
Inoculation of potato plants of different cultivars with R. solanacearum showed low levels of chlorophyll pigments in the leaves which indicated a process of senescence. During senescence, leaf cells undergo drastic metabolic degeneration of cellular structures, starting with chlorophyll catabolism as well as protein and RNA degradation, with loss of photosynthetic activity and chlorophyll content, which were greater in susceptible potato cvs. compared to the tolerant potato cvs. [19] . It was obvious that rate of chlorophyll loss due to infection with R. solanacearum was negatively correlated with tolerance of potato cvs. to R. solanacearum. Increased plant defense-related enzyme activities were observed in potato cultivars tolerant to the infection with bacterial pathogen compared to the susceptible ones [20] - [23] . The oxidative enzymes such as POD and PPO, can catalyze the formation of lignin and other oxidative phenols, and contribute in formation of defense barriers by changing the cell structure defense system get actuated against pathogens [24] . The resistance induced by systemic acquired resistance (SAR) is generally effective against a broad range of pathogens and this type of resistance is associated with an increase in the activity of POD [25] , PAL [26] and PPO in the plant [26] .
In present study, the relatively highly resistant cultivar (Nicola) showed an increase in POD activity in which it was noticed initially and reached its peak 0.488 min−1∙g−1 at 12 h, and kept at the higher level up to 72 h after inoculation. On the contrary, in the susceptible, cv. Spunta, the POD activity peak was as low as 0.226 min−1∙g−1 and was recognized later at 48 h, after inoculation. Potato, cv. Kara, however, of the intermediate resistance to R. solanacearum, showed intermediate POD activity for the peak value and its time. In tomato, POD is one of the enzymes believed to catalyse the last step in the lignification pathways. The reinforcement of the plant cell wall by phenolics and lignin increases plant resistance to wall-degrading enzymes produced by pathogens, and acts as a mechanical barrier to toxin ingress and to physical penetration toward the protoplast [28] .
POD activity in plants can increase in response to a variety of stresses including biotic stress, indicating that POD activities involved in defense against attack by pathogens [29] . In the present study, we reported the quick response of POD when potato inoculated with R. solanacearum, indicating a possible role of the enzyme during pathogen infection and host-resistance. The fact that POD activity was higher in resistant and moderately resistant cultivars than in susceptible cultivar indicates that POD might have played a specific role in triggering the development of host resistance. Our findings are similar with the results of the earlier studies of Chittoor et al. [30] on rice in case of Xanthomonas oryzae pv. oryzae and Vanitha & Umesha [31] on tomato in case of R. solanacearum path-systems. In the relatively highly resistant cultivars, the activity of POD was higher when compared to the un-inoculated plants and also with the susceptible and highly susceptible cultivars.
Under stress conditions, the enhanced POD activity in the intercellular spaces, stimulating cell wall stiffening, probably reduces cell growth which might represent a mechanical adaption to adverse conditions [32] . Meanwhile, an enhanced POD activity was demonstrated during various pathogenesis systems [29] .
Polyphenol oxidase, a copper containing enzyme, oxidizes phenolics to highly toxic quinines and is involved in the terminal oxidation of diseased plant tissue and is attributed for its role in disease resistance [33] .
Concerning PPO activity, in resistant cultivar (Nicola) the peak of activity of PPO was the highest (0.445 min−1∙g−1) early at 24 h, after inoculation with R. solanacearum. However, in susceptible cultivar (Spunta), maximum activity was lower (0.364 min−1∙g−1) and was recognized as later at 48 h after inoculation. Meanwhile, cv. Kara showed intermediate values. PPO is a copper-containing enzyme known to be involved in resistance against R. solanacearum in resistant tomato cultivars [34] . Besides, overexpression of PPO in transgenic tomato plants enhanced their resistance to Pseudomonas syringae, another bacterial pathogen of tomato [35] . Also, there are reports that PPO activities increased in resistant tomato cultivars more than those in susceptible and highly susceptible cultivars after inoculation with Xanthomonas axonopodis pv. vesicatoria [36] . Increased PPO activity contributed to disease resistance due to its property to oxidize phenolic compounds to more toxic quinines which badly affect pathogenic micro-organisms [37] .
On the contrary, CAT activity, in the highly susceptible cv. Spunta, was always higher than its level in both, the resistant cultivar (Nicola) and moderately resistant one (Kara). CAT is the key H2O2 detoxifying enzyme in plant which keeps the balance of the active oxygen species (AOS), such as H2O2 level during plant defense. H2O2 is associated with hypersensitive response (HR) during systemic acquired resistance. Higher concentrations of H2O2 in resistant than in susceptible tomato cultivars have been reported in tomato-Ralstonia interactions [7] . Another study reported that the restriction of R. solanacearum growth could be due to the antimicrobial activity of H2O2, which is strongly increased around bacterial cells. Consequently, the high CAT susceptible cultivars restrict H2O2 action against R. solanacaerum.
To investigate the association of gene expression of potato cultivars and resistance to R. solanacaerum. The real-time qPCR technique was used to evaluate changes in the transcription levels of three genes peroxidase, polyphenol oxidase and catalase. At zero time, the relatively highly resistant potato cultivar, Nicola, showed the highest values of gene expression for both POD and PPO. However, the susceptible potato cultivars, Spunta, showed the lowest values, while, the intermediate resistance potato cultivars, Kara, exhibited intermediate values.
On the contrary, the CAT, gene expression was the highest in the susceptible potato, cv. Spunta, and was the lowest in the relatively highly resistant potato, cv. Nicola, while, was of intermediate values in cv. Kara (intermediate resistance).
Meanwhile, gene expression reached its peak (4.2) for POD in cv. Nicola after 12 hours while, it was at 48 hours for Spunta and at low peak of 2.0. Intermediate values were recognized for cv. Kara. Similar trend was revealed for PPO gene expression.
However, an opposite trend was revealed for CAT where cv. Nicola (relatively highly resistant) showed the lowest values while, cv. Spunta (susceptible) showed the highest values. Also, cv. Kara showed intermediate values. These results were in harmony with findings of Navodit & Prabir [38] . Peroxidase gene expression in tomato was shown to be induced differentially in resistant and susceptible lines by elicitors of the fungal pathogen Verticillium albo-atrum [39] .
Also, catalase expression and activity change during plant-pathogen interactions and a decrease in plant catalase activity occurs in resistant plants in response to attempted infection by viruses [40] . It is thought that this allows H2O2 to accumulate, resulting in antimicrobial activity through strengthening of the plant cell wall, activation of defense genes, hypersensitive cell death and a subsequent halt to pathogen infection [41] . In contrast, in susceptible hosts increases in host catalase activity have been observed by Havelda & Maule, [42] Kuzniak & Sklodowska, [43] and Pompe-Novaka et al., [44] , exogenously applied catalase can result in decreased hypersensitive cell death [45] and increased penetration by pathogens of normally resistant hosts (Borden & Higgins, [46] and Abel et al., [47] .
5. Conclusions
Finally, Peroxidase (POD), Polyphenol oxidase (PPO) and Catalase (CAT) gene expressions and enzymes activities can be used as molecular and biochemical markers to reveal the resistance or susceptibility nature of potato cultivars against bacterial wilt disease of potato caused by R. solanacaerum.
Cite this paper
Eman El-Argawy,Ibrahim A. Adss, (2016) Quantitative Gene Expression of Peroxidase, Polyphenoloxidase and Catalase as Molecular Markers for Resistance against Ralstonia solanacearum. American Journal of Molecular Biology,06,88-100. doi: 10.4236/ajmb.2016.62010
References
- 1. FAO, STAT (2013) FAOSTAT Database. Food and Agriculture Organization of the United Nations, Rome, Italy.
http://faostat.fao.org - 2. Milling, A., Babujee, L. and Allen, C. (2011) Ralstonia solanacearum Extracellular Polysaccharide Is a Specific Elicitor of Defense Responses in Wilt Resistant Tomato Plants. PLoS ONE, 6, Article ID: e15853.
http://dx.doi.org/10.1371/journal.pone.0015853 - 3. Aliye, N., Fininsa, C. and Hiskias, Y. (2008) Evaluation of Rhizosphere Bacterial Antagonists for Their Potential to Bioprotect Potato (Solanum tuberosum) against bacterial wilt (Ralstonia solanacearum). Biological Control, 47, 282-288.
http://dx.doi.org/10.1016/j.biocontrol.2008.09.003 - 4. Chandrashekara, K.N., Prasanna, M.K. and Saroja, S. (2012) Aggressiveness of Ralstonia solanacearum Isolates on Tomato. Journal of Experimental Sciences, 3, 5-9.
- 5. Adss, I.A. (2014) Different Gene Expression of Polygalacturonase (pehC) and Its Relationship to the Pathogenicity of Different R. solanacearum Isolates. Journal of Agriculture & Environmental Sciences Damanhur University, Egypt, 31, 48-62.
- 6. Soares, A.G., Trugob, L.C., Botrela, N. and Silva, L.F. (2005) Reduction of Internal Browning of Pineapple Fruit (Ananascomosus L.) by Preharvest Soil Application of Potassium. Postharvest Biology and Technology, 35, 201-207.
http://dx.doi.org/10.1016/j.postharvbio.2004.07.005 - 7. Mandal, S., Das, R.K. and Mishra, S. (2011) Differential Occurrence of Oxidative Burst and Antioxidative Mechanism in Compatible and Incompatible Interactions of Tomato and Ralstonia solanacearum. Plant Physiology and Biochemistry, 49, 117-123.
http://dx.doi.org/10.1016/j.plaphy.2010.10.006 - 8. Nolan, T., Hands, R.E. and Bustin, S.A. (2006) Quantification of mRNA Using Real-Time RT-PCR. Nature Protocols, 1, 1559-1582.
http://dx.doi.org/10.1038/nprot.2006.236 - 9. Deihimi, T., Niazi, A. and Ebrahimie, E. (2013) Identification and Expression Analysis of TLPs as Candidate Genes Promoting the Responses to Both Biotic and Abiotic Stresses in Wheat. Plant Omics, 6, 107-115.
- 10. Prior, P. and Steva, H. (1990) Characteristics of Strains of Pseudomonas solanacearum from the French West Indies. Plant Disease, 74, 13-17.
http://dx.doi.org/10.1094/PD-74-0013 - 11. Tans-Kersten, J., Huang, H. and Allen, C. (2001) Ralstonia solanacearum Needs Motility for Invasive Virulence on Tomato. Journal of Bacteriology, 183, 3597-3605.
http://dx.doi.org/10.1128/JB.183.12.3597-3605.2001 - 12. Wintermans, J.E.G. and De Mots, A. (1965) Spectrophotometric Characteristics of Chlorophyll a and b and Their Phaeophytins in Ethanol. Biochimica et Biophysica Acta, 109, 448-453.
http://dx.doi.org/10.1016/0926-6585(65)90170-6 - 13. Hammerschmidt, R., Nuckles, E. and Kuc, J. (1982) Association of Enhanced Peroxidase Activity with Induced Systemic Resistance of Cucumber to Colletotrichum lagenarium. Physiologia Plantarum, 20, 73-80.
http://dx.doi.org/10.1016/0048-4059(82)90025-x - 14. Mayer, A.M., Harel, E. and Shaul, R.B. (1965) Assay of Catechol Oxidase: A Critical Comparison of Methods. Phytochemistry, 5, 783-789.
http://dx.doi.org/10.1016/S0031-9422(00)83660-2 - 15. Chandlee, M. and Scandalios, J.G. (1984) Analysis of Variants Affecting the Catalase Developmental Program in Maize Scutellum. Theoretical and Applied Genetics, 9, 71-77.
http://dx.doi.org/10.1007/bf00262543 - 16. Albayrak, G. and Arican, E. (2004) Amplification of Specific Genes by Using RT-PCR Technique in Plants. Biotechnology & Biotechnological Equipment, 18, 15-18.
http://dx.doi.org/10.1080/13102818.2004.10819223 - 17. Rasmussen, R. (2001) Quantification on the Light Cycler. In: Meuer, S., Wittwer, C. and Nakagawara, K., Eds., Rapid Cycle Real-Time PCR, Methods and Applications, Springer Press, Heidelberg, 21-34.
- 18. SAS Institute (2000) SAS Users Guide, Version 8.1. SAS Institute, Cary, 286-288.
- 19. Hörtensteiner, S. and Feller, U. (2002) Nitrogen Metabolism and Remobilization during Senescence. Journal of Experimental Botany, 53, 927-937.
http://dx.doi.org/10.1093/jexbot/53.370.927 - 20. Haasan, M.A.E. and Buchenauer, H. (2007) Induction of Resistance to Fire Blight in Apple by Acibenzolar-S-methyl and DL-3-aminobutryic Acid. Journal of Plant Diseases and Protection, 114, 151-158.
- 21. Mustafa, H.S.A. and Alawami, A.M.Y. (2012) Susceptibility of Newly Introduced Potato Cultivars to Libya to Infection with Bacterial Soft Rot and the Associated Physiological Changes. Journal of Agricultural Science and Technology, 2, 976-982.
- 22. Ngadze, E., Icishahayo, D., Coutinho, T.A. and Van der Waals, J.E. (2012) Role of Polyphenol Oxidase, Peroxidase, Phenylalanine Ammonia Lyase, Chlorogenic Acid and Total Soluble Phenols in Resistance of Potatoes to Soft Rot. Plant Disease, 96, 186-192.
http://dx.doi.org/10.1094/PDIS-02-11-0149 - 23. Nisha, S., Revathi, K., Chandrasekaran, R., Kirubakaran, S.A., Sathish-Narayanan, S., Stout, M.J. and Senthil-Nathan, S. (2012) Effect of Plant Compounds on Induced Activities of Defense-Related Enzymes and Pathogenesis Related Protein in Bacterial Blight Disease Susceptible Rice Plant. Physiological and Molecular Plant Pathology, 80, 1-9.
http://dx.doi.org/10.1016/j.pmpp.2012.07.001 - 24. Thilagavathi, R., Saravanakumar, D., Ragupathi, N. and Samiyappan, R. (2007) A Combination of Bio-Control Agents Improves the Management of Dry Root Rot (Macrophomina phaseolina) in Greengram. Phytopathologia Mediterranea, 46, 157-167.
- 25. Van Loon, L.C. (1997) Induced Resistance in Plants and the Role of Pathogenesis-Related Proteins. European Journal of Plant Pathology, 103, 753-765.
http://dx.doi.org/10.1023/A:1008638109140 - 26. Raj, S.N., Sarosh, B.R. and Shetty, H.S. (2006) Induction and Accumulation of Polyphenol Oxidase Activities as Implicated in Development of Resistance against Pearl Millet Downy Mildew Disease. Functional Plant Biology, 33, 563-571.
http://dx.doi.org/10.1071/FP06003 - 27. Mayer, A.M. and Staphes, R.C. (2002) Laccase: New Functions for an Old Enzyme. Phytochemistry, 60, 551-565.
http://dx.doi.org/10.1016/S0031-9422(02)00171-1 - 28. Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic Compounds and Their Role in Disease Resistance. Annual Review of Phytopathology, 30, 369-389.
http://dx.doi.org/10.1146/annurev.py.30.090192.002101 - 29. Kartashova, E.R., Rudenskaya, G.N. and Yurina, E.V. (2000) Plant Peroxidase Polyfunctionality and Their Application in Practice. Journal of Agricultural Biology, 1, 63-70. (In Russian)
- 30. Chittoor, J.M., Leach, H.E. and White, F.F. (1997) Differential Induction of Peroxidase Gene Family during Infection of Rice by Xanthomonas oryzae pv. oryzae. Molecular Plant-Microbe Interactions Journal, 10, 861-871.
http://dx.doi.org/10.1094/MPMI.1997.10.7.861 - 31. Vanitha, S.C. and Umesha, S. (2008) Variations in Defense Related Enzyme Activities in Tomato during the Infection with Bacterial Wilt Pathogen. Journal of Plant Interactions, 3, 245-253.
http://dx.doi.org/10.1080/17429140802032863 - 32. Ranieri, A., Nali, G.D. and Urso, G. (1995) Peroxidase Activity in Cucurbita pepo L. Leaves Exposed to Ozone. Agricoltura Mediterranea, Special Volume, 47-54.
- 33. Barilli, E., Prats, E. and Rubiales, D. (2010) Benzothiadiazole and BABA Improve Resistance to Uromyces pisi (Pers.) Wint. in Pisum sativum L. with an Enhancement of Enzymatic Activities and Total Phenolic Content. European Journal of Plant Pathology, 128, 483-493.
- 34. Vanitha, S.C., Niranjana, S.R. and Umesha, S. (2009) Role of Phenylalanine Ammonia Lyase and Polyphenol Oxidase in Host Resistance to Bacterial Wilt of Tomato. Journal of Phytopathology, 157, 552-557.
http://dx.doi.org/10.1111/j.1439-0434.2008.01526.x - 35. Li, L. and Steffens, J.C. (2002) Overexpression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance. Planta, 215, 239-247.
http://dx.doi.org/10.1007/s00425-002-0750-4 - 36. Kavitha, R. and Umesha, S. (2008) Regulation of Defense-Related Enzymes Associated with Bacterial Spot Resistance in Tomato. Phytoparasitica, 36, 144-159.
http://dx.doi.org/10.1007/BF02981327 - 37. Vinale, F., Sivasithamparam, K., Ghisalberti, E.L., Marra, R., Woo, S.L. and Lorito, M. (2008) Trichoderma-Plant-Pathogen Interactions. Soil Biology and Biochemistry, 40, 1-10.
http://dx.doi.org/10.1016/j.soilbio.2007.07.002 - 38. Navodit, G. and Prabir, K.P. (2014) Induction of Systemic Resistance in Tomato by Fruit Extracts of Azadirachta indica. Reviews of Literature, 2, 1-27.
- 39. Mohan, R. and Kolattukudy, P.E. (1990) Differential Activation of Expression of a Suberization-Associated Anionic Peroxidase Gene in Near-Isogenic Resistant and Susceptible Tomato Lines by Elicitors of Verticillium alboatrum. Plant Physiology, 92, 276-280.
http://dx.doi.org/10.1104/pp.92.1.276 - 40. Yi, S.Y., Yu, S.H. and Choi, D. (2003) Involvement of Hydrogen Peroxide in Repression of Catalase in TMV-Infected Resistant Tobacco. Molecules and Cells, 15, 364-369.
- 41. Lamb, C. and Dixon, R.A. (1997) The Oxidative Burst in Plant Disease Resistance. Annual Review of Plant Physiology and Plant Molecular Biology, 48, 251-275.
http://dx.doi.org/10.1146/annurev.arplant.48.1.251 - 42. Havelda, Z. and Maule, A.J. (2000) Complex Spatial Responses to Cucumber Mosaic Virus Infection in Susceptible Cucurbita pepo Cotyledons. Plant Cell, 12, 1975-1985.
http://dx.doi.org/10.1105/tpc.12.10.1975 - 43. Kuzniak, E. and Sklodowska, M. (2005) Fungal Pathogen-Induced Changes in the Antioxidant Systems of Leaf Peroxisomes from Infected Tomato Plants. Planta, 222, 192-200.
http://dx.doi.org/10.1007/s00425-005-1514-8 - 44. Pompe-Novak, M., Gruden, K. and Baebler, S. (2006) Potato Virus Y Induced Changes in the Gene Expression of Potato (Solanum tuberosum L.). Physiological and Molecular Plant Pathology, 67, 237-247.
http://dx.doi.org/10.1016/j.pmpp.2006.02.005 - 45. Able, A.J., Guest, D.I. and Sutherland, M.W. (2000) Hydrogen Peroxide Yields during the Incompatible Interaction of Tobacco Suspension Cells Inoculated with Phytophthora nicotianae. Plant Physiology, 124, 899-910.
http://dx.doi.org/10.1104/pp.124.2.899 - 46. Borden, S. and Higgins, V.J. (2002) Hydrogen Peroxide Plays a Critical Role in the Defense Response of Tomato to Cladosporium fulvum. Physiological and Molecular Plant Pathology, 61, 227-236.
http://dx.doi.org/10.1006/pmpp.2002.0435 - 47. Abel, A.J., Sutherland, M.W. and Guest, D.I. (2003) Production of Reactive Oxygen Species during Non-Specific Elicitation, Non-Host Resistance and Field Resistance Expression in Cultures of Tobacco Cells. Functional Plant Biology, 30, 91-99.
http://dx.doi.org/10.1071/FP02123
上一篇:TRPC3 and TRPC6 Contribute to 下一篇:Constitutive and Secretory Exp
最新文章NEWS
- TRPC3 and TRPC6 Contribute to the Pathogenesis of Hypertension
- Quantitative Gene Expression of Peroxidase, Polyphenoloxidase and Catalase as Molecular Markers for
- Constitutive and Secretory Expression of the AiiA in Pichia pastoris Inhibits Amorphophallus konjac
- Association of XbaI GLUT1 Polymorphism with Susceptibility to Type 2 Diabetes Mellitus and Diabetic
- Comparison of African Swine Fever Virus Prevalence in Nigerian Indigenous Pig, Its Hybrid and Backcr
- Isolation and Characterization of Hydrocarbon Utilizing Yeast (HUY) Isolates from Palm Wine
- Haematological Traits of Nigerian Indigenous Pig and Its Hybrid (50% Large White × 50 NIP) at Post W
- Synapsin Polymorphisms Could Be Correlated with Stroop Simple Reaction Time Scores
推荐期刊Tui Jian
- Chinese Journal of Integrative Medicine
- Journal of Genetics and Genomics
- Journal of Bionic Engineering
- Pedosphere
- Chinese Journal of Structural Chemistry
- Nuclear Science and Techniques
- 《传媒》
- 《中学生报》教研周刊
热点文章HOT
- Isolation and Characterization of Hydrocarbon Utilizing Yeast (HUY) Isolates from Palm Wine
- Haematological Traits of Nigerian Indigenous Pig and Its Hybrid (50% Large White × 50 NIP) at Post W
- Updating Genetics Polymorphisms of Non-Syndromic Clefts Lip-Palates
- Molecular Cloning and Expression Analysis of PgLAC in Pomegranate
- Genetic Diversity of Maize Accessions (Zea mays L.) Cultivated from Benin Using Microsatellites Mark
- Phosphorylation on TRPV4 Serine Residue 824 Enhances Its Association with PGM1
- Molecular Cloning and Tissue Distribution of Troponin C from the Japanese Pearl Oyster, Pinctada fuc
- Genetic Polymorphisms of Nervous System Development and the Risk of Posttraumatic Stress Disorder

