Characterization of Yeasts Capable of Neutralizing Acidic Media from Natural Neutral Environments
Vol.07No.12(2017), Article ID:81331,11 pages
10.4236/aim.2017.712068
Shintaro Nagaoka, Takako Kobayashi, Yuko Kajiwara, Masahiko Okai, Masami Ishida, Naoto Urano
Laboratory of Marine Biochemistry, Department of Ocean Sciences, Tokyo University of Marine Science and Technology, Tokyo, Japan
Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: November 6, 2017; Accepted: December 24, 2017; Published: December 27, 2017
ABSTRACT
Acid-tolerant yeasts often inhabit extremely acidic environments: mine drainages, hot springs, and even fermented foods. Some of them also possess the ability to neutralize acidic media. However, the examples of these yeasts that are already known were isolated from acidic environments. In this study, the isolation of acid-tolerant yeasts from natural neutral aquatic environments and the identification of yeasts able to neutralize an acidic medium (acid-neutralizing yeast) in Japan were carried out. Various kinds of acid-tolerant and acid-neutralizing yeasts were obtained. In a neutralizing test using an acidic casamino acid solution adjusted to a pH of 4.0 with sulfuric acid, the obtained acid-neutralizing yeasts elevated the pH to approximately 7.0, and their neutralizing abilities were similar to those of previously reported yeasts that had been isolated from acidic environments. These results showed that acid-tolerant yeasts and acid-neutralizing yeasts exist widely in neutral environments, and little difference was found in the neutralizing abilities of yeasts obtained from neutral environments in comparison to those obtained from acidic environments.
Keywords:
Acid-Tolerant Yeast, Acidic Environment, Hydrosphere, Neutralization, Yeast Species

1. Introduction
Extremophiles are organisms that thrive in extreme environments such as extremely high or low temperature, high pressure, high salinity, and high or low pH [1] . For instance, Thermus aquaticus, a bacterium isolated from Mushroom Spring in Yellowstone National Park, flourishes at 70˚C [2] . Halobacteroides halobius is a bacterium isolated from Dead Sea sediment which requires high NaCl concentrations (between 1.4 and 2.8 mol/l) for growth [3] , and Sulfolobus tokodaii is an archaeon that thrives at pH 2.5 - 3, and furthermore at 80˚C [4] . Many species of archaea and bacteria are known as extremophiles, but the taxonomic range spans all three domains [1] .
In this study, we focused on eukaryotes that can inhabit acidic environments. Some previous studies reported that a variety of eukaryotes exist in extreme acidic environments, not only prokaryotes. For instance, Klebsormidium and Dunaliella, which are algae, have been isolated from acidic rivers [5] . Additionally, several types of yeast present in acidic environments have also been extensively investigated, e.g., Rhodotorula glutinis [6] and Candida multosa [7] . Moreover, we found that both Candida fluviatilis CeA16 and Cryptococcous sp. T1 were acid-tolerant yeasts and had the ability to neutralize acidic media by decomposing amino acids to release ammonium ions [8] [9] . While the mechanisms of acid-tolerant bacteria have been well investigated [10] , the mechanisms by which yeasts neutralize acidic environments remain unclear.
The yeasts already known to be capable of thriving in and neutralizing acidic environments were isolated only from extreme acidic environments. To our knowledge, there has been no report of such yeasts being isolated from a neutral natural environment. In this study, we identified various types of acid-tolerant yeasts that possess the ability to neutralize an acidic medium in neutral aquatic environments and characterized their abilities.
2. Materials and Methods
2.1. Collection of Environmental Samples
From April 2016 to June 2017, we collected water and sediment samples from various water environments, namely two locations around Tokyo University of Marine Science and Technology, in Tokyo, and 12 locations in Yokohama, Kanagawa prefecture, Japan. All the samples were collected in sterile tubes, and the pH value of each sample was measured. As for the sediment sample, a small amount of distilled water was added and the sample was vortexed, then the pH value was measured. The samples were carried and stored at low temperatures in our laboratory at Tokyo University of Marine Science and Technology.
2.2. Medium Culture
For cultivation, we used an R2A medium and a YPD medium. The R2A medium consisted of 0.1% yeast extract (Becton Dickinson, Franklin Lakes, NJ, USA), 0.1% proteose peptone (Becton Dickinson), 0.1% casamino acid (Nihon Pharmaceutical, Tokyo), 0.1% D-(+)-glucose, 0.1% soluble starch (Kokusan Chemicals, Tokyo), 0.06% sodium pyruvate, 0.03% K2HPO4, and 0.005% MgSO4∙7H2O. The YPD medium consisted of 1.0% yeast extract (Becton Dickinson), 2.0% proteose peptone (Becton Dickinson), and 2.0% D-(+)-glucose. We prepared neutral media or acidic media adjusted to pH 3.0 with sulfuric acid, and the solid media were constructed by adding 1.2% gellan gum (Kanto Chemical, Tokyo) to both R2A and YPD liquid media. The neutral media contained 0.01% chloramphenicol to prevent bacteria growth.
2.3. Isolation of Acid-Tolerance Yeast Strains
For the isolation of yeasts, we first used both R2A (1) medium (neutral R2A solid medium) and YPD (1) medium (neutral YPD solid medium). The water samples were filtered with a 0.45-µm FTFE membrane-filter (Advantec, Tokyo), and microorganisms were trapped on the filter. They were dispersed into the portion of the filtrate; thus, we obtained about a 100-fold-concentrated population of the microorganisms in the water samples. The sediment samples were diluted to 10-fold with physiological saline (0.8%w/v NaCl). A 200 µl volume of each preparation was spread on the R2A and YPD media and incubated at 25˚C. After several days, we picked up growing yeast-like colonies: i.e., we attached the colonies to the tip of a needle and transferred them to a new medium.
To isolate acid-tolerant yeasts, the obtained colonies were inoculated into acidic R2A (2) medium (R2A solid medium adjusted to pH 3.0 with sulfuric acid) or acidic YPD (2) medium (YPD solid medium adjusted to pH 3.0 with sulfuric acid) and then incubated at 25˚C.
Colonies formed on acidic solid medium were removed and inoculated into R2A (3) medium (R2A plate medium adjusted to pH 3.0 with sulfuric acid which contained 0.02% bromocresol purple, which is yellow in acidic conditions and purple in neutral and basic conditions). After several days’ incubation at 25˚C, we separated the yeasts that were capable of neutralizing acid (hereafter referred to as acid-neutralizing yeasts) by observing the change of the medium color from yellow to purple.
2.4. Yeast Identification
The yeasts inoculated on YPD (1) medium formed colonies that were over 2 cm in diameter (giant colonies) after several days’ incubation at 25˚C. First, we distinguished yeast species among the isolates by observing the appearance of giant colonies with the naked eye and observing the cell morphologies under a microscope. Second, with the yeasts thus obtained, we carried out the gene identification. The 26S rRNA genes of the yeasts were amplified by polymerase chain reaction (PCR) using the forward primer NL-1 (5'-GCATATCAATAAGCGGAGGAAAAG-3'), and the reverse primer NL-4 (5'-GGTCCGTGTTTCAAGACGG-3'), and Premix Ex Taq (Takara Bio, Shiga, Japan). The PCR reaction program was performed in a TaKaRa PCR Thermal Cycler Dice TP600 with an initial denaturation at 95˚C for 5 min, followed by 25 cycles of denaturation at 94˚C for 30 s, annealing at 56˚C for 30 s, extension at 72˚C for 1 min and final extension at 72˚C for 4 min. The D1/D2 domain sequences of the 26S rRNA genes in the yeasts were deposited in DNA Data Bank of Japan (DDBJ), European Molecular Biology Laboratory (EMBL), and GenBank.
2.5. Neutralization of Casamino Acid Solution at pH 4.0
To investigate the neutralization ability of the yeasts, we carried out a neutralization test using the isolated acid-neutralizing yeasts. The yeasts were precultured at 25˚C for 48 h in 10 mL of R2A liquid medium at pH 4.0 with shaking. The yeast pellet obtained by the centrifugation at 3000 rpm was washed three times with saline adjusted to pH 4.0 with sulfuric acid. The washed pellet was added to 10 mL of 0.5% (w/v) casamino acid solution adjusted to pH 4.0 with sulfuric acid, and incubated at 25˚C for 72 h with shaking. We prepared four same 10 ml samples. One of them was picked up and centrifuged at 3000 rpm, and the pH value of the supernatant was then measured every 24 h.
3. Results
3.1. Yeast Isolation and Identification
A total of 177 yeast-like strains were obtained from all water and sediment samples. Forty-six strains changed the color of the R2A (2) medium containing a pH indicator from yellow to purple (an example of the change of color is shown in Figure 1). As a result of the microscopic observation, 12 bacteria-like strains were excluded, and 34 yeast-like strains were finally isolated as acid-neutralizing yeasts. Figure 2 shows micrograph examples of acid-neutralizing yeasts. With these 34 strains, we performed giant colony observation to distinguish their species. After that, when species that greatly resembled other species were excluded, 26 strains were identified, and the results are shown in Table 1 and Table 2. As a result, four strains (r-w2, r-w3, r-w4, and fi-m10) showed sequences identical
 (a)
(a)  (b)
(b)
Figure 1. An example of the change in the R2A plate medium color. The neutralization by strain sm-w37 (P. antarctica) inoculated into the acidic plate medium (a) elevated the surrounding pH, and the color of the medium changed to purple (b) after 7 days incubation at 25˚C.
 (a)
(a)  (b)
(b)
Figure 2. Micrograph of acid-neutralizing yeasts. (a) is strain fi-m10 (identified as A. pullulans); (b) is strain sm-w39 (identified as P. tsukubaensis). After each strain was incubated in acidic R2A liquid medium adjusted to pH 3.0 with sulfuric acid at 25˚C for 48 h with shaking, a portion of the medium was observed with an optical microscope.
Table 1. Isolated acid-neutralizing yeast strains and pH and temperature of sampling areas.
aThe total number of colonies isolated from samples. bObtained strains that were acid-neutralizing yeasts.
Table 2. Identification of acid-neutralizing yeasts.
to Aureobasidium pullulans. The strains h-m7, so-w34, and n-w33 were found to be Candida parapsilosis, C. cylindracea, and C. oleophila, respectively, and two strains (h-m8 and om-w46) were found to be Candida sp. Two strains (ko-w20, om-w42) were Cryptococcus sp. and two strains (m-m25 and n-w28) were Cryptococcus flavescens. Two strains (si-w12 and n-w29) were Rhodotorula sp. Strain mi-w17 was Hannaella pagnoccae, and strain sm-w38 was H. coprosmae. Strain sm-w37 was found to be Pseudozyma antarctica, and sm-w39 was P. tsukubaensis. Strains mi-w16, si-w13, om-41, om-w44, sm-w40, ks-w31, and mr-w1 were Filobasidium magnum, Leucosporidium golubevii, Auriculibuller sp., Papiliotrema flavescens, Microbotryozyma collariae, Bullera alba, and Meyerozyma guilliermondii, respectively. The phylogenetic tree of these species was constructed on the 26s rRNA sequences by the maximum likelihood algorithm in MEGA version 6.06, as shown in Figure 3.
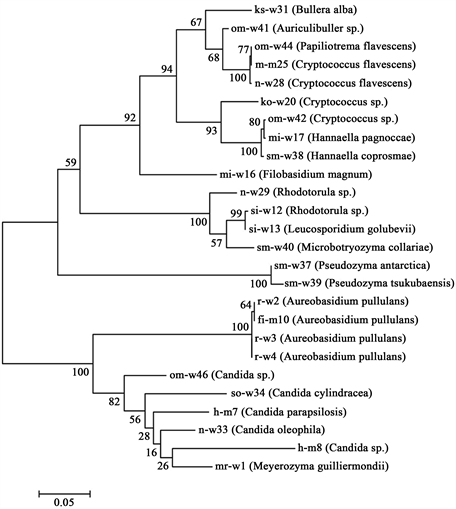
Figure 3. Phylogenetic tree of isolated acid-neutralizing yeasts. The phylogenetic tree was constructed in MEGA version 6.06 using the maximum likelihood method with a 1000 replicate bootstrap resampling.
3.2. Neutralization of Casamino Acid Solution
Among the 26 identified strains, 10 strains (h-m7, ko-w20, ks-w31, m-m25, mi-w16, mi-w17, n-w29, r-w2, r-w3, and si-w13) changed the color of R2A (2) medium faster than the others. With these 10 strains, we performed neutralizing tests using casamino acid solution adjusted to pH 4.0 with sulfuric acid, and the results are shown in Figure 4. All strains elevated the pH from 4.0 to approximately 7.5 after 24 h, and from 24 h to 72 h the pH increased gradually up to approximately 8.0.
4. Discussion
In this study, we isolated 26 acid-neutralizing yeasts which exhibited acid tolerance and the ability to neutralize acidic R2A (2) medium from 14 natural neutral aquatic locations in Tokyo and Yokohama, Kanagawa prefecture, Japan. These 26 isolates were identified. Using 10 strains that rapidly changed the color of R2A (2) medium containing bromocresol purple from yellow to purple, we

Figure 4. Neutralization of casamino acid solutions by the 10 isolated acid-neutralizing yeasts. Each strain was precultured at 25˚C for 48 h in 10 mL of R2A liquid medium at pH 4.0 with shaking. After the yeast pellet was obtained by centrifugation and washed three times, the washed pellet was added to 10 mL of 0.5% (w/v) casamino acid solution at pH 4.0, and incubated at 25˚C for 72 h with shaking. Four same samples were prepared, and one of them was collected every 24 h. After centrifugation at 3000 rpm, the pH value of the supernatant was measured.
carried out neutralizing tests using 0.5% (w/v) casamino acid solution adjusted to pH 4.0 with sulfuric acid.
In previous reports, yeast strains identified as C. fluviatilis [11] [12] , C. intermedia [12] , and R. glutinis [6] were isolated from various extreme acidic environments, and some of them had the ability to neutralize acidic media (pH 2.5 - 3.0) [6] [12] . However, there have been no reports of the isolation of acid-neutralizing yeast from a neutral aquatic environment.
Table 1 and Table 2 show the results of the identification of 26 isolated acid-neutralizing yeasts and also show the pH and temperature of each sampling area. Various species were obtained, and it is assumed that acid-neutralizing yeasts exist not only in acidic environments but also in neutral environments. Figure 3 shows the phylogenetic tree of isolated acid-neutralizing yeasts. Many strains were identified as Cryptococcus sp. and Candida sp. Furthermore, the figure shows wide variation in species in the identification results. In particular, the 26S rRNA sequence of strain ko-w20 (isolated from Kodomo Shizen Park, Yokohama) showed 100% identity with that of Cryptococcus sp. strain T1, which is an acid-neutralizing yeast isolated from Lake Tazawa, an acidic lake in Japan [9] . Furthermore, 22 strains were obtained from water samples and four strains were isolated from sediments; this indicates that water environments are the major habitat of acid-neutralizing yeasts.
As a result of the neutralizing test, 10 strains (strain m-m25 was from sediment, while the nine other strains were from water) elevated the pH from 4.0 to approximately 8.0 after 72 h (Figure 4). In the acid-tolerant yeast isolation step (as mentioned above in Section 2.3), we used R2A (2) medium, whose pH is 3.0, but we used pH 4.0 casamino acid solution in the neutralizing test. We did so because some strains had not neutralized pH 3.0 casamino acid solution (data not shown), so for a comparison of the neutralizing ability we chose pH 4.0 solution. There were no significant differences in the neutralizing ability among these strains, and the level of pH increase was similar to the result of the same test using C. fluviatilis strain CeA16, which was isolated from an extremely acidic river and was found to be capable of neutralizing acid in prior research [8] . These results might indicate that the ability of acid-neutralizing yeasts to neutralize acidic casamino acid solution is approximately equal regardless of the pH value of the location they were isolated from. In the same research [8] , the neutralizing test with C. fluviatilis strain CeA16 showed a decrease in the amino acid concentration and an increase in the ammonium ion concentration in an acidic casamino acid solution as the pH value increased. As for the present study, the amino acid and ammonium ion concentrations were not measured; however, we inferred that the same type of phenomena might have occurred, specifically an increase in the ammonium ion concentration. The results of previous study [8] and the present study suggest that acid-neutralizing yeasts decompose amino acids into ammonium ions by an enzyme reaction, and then neutralize the acidic condition (Figure 5).

Figure 5. Hypothesized neutralization mechanism of acid-neutralizing yeast. The yeast takes free amino acid into the cell, produces ammonium ions by an enzyme reaction, and then releases the ammonium ions out of the cell to neutralize the external acidic condition.
5. Conclusion
In conclusion, we isolated 26 strains of neutralizing yeast from neutral water environments. Of these, 10 strains elevated the pH of acidic casamino acid solution from 4.0 to approximately 8.0 in 72 h, and there were no significant differences in neutralizing ability among them. This study indicates that neutralizing yeasts exist widely in the neutral environment. The results presented in this study may further our understanding of the acid tolerance and neutralizing ability of yeasts.
Acknowledgements
This work was supported in part by a Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research (Grant No. 16K07868).
Cite this paper
Nagaoka. S., Kobayashi, T., Kajiwara, Y., Okai, M., Ishida, M. and Urano, N. (2017) Characterization of Yeasts Capable of Neutralizing Acidic Media from Natural Neutral Environments. Advances in Microbiology, 7, 887-897. https://doi.org/10.4236/aim.2017.712068
References
- 1. Rothschild, L.J. and Mancinelli, R.L. (2001) Life in Extreme Environments. Nature, 409, 1092-1101. https://doi.org/10.1038/35059215
- 2. Brock, T.D. and Freeze, H. (1969) Thermus aquaticus gen. n. and sp. n., a Nonsporulating Extreme Thermophile. Journal of Bacteriology, 98, 289–297.
- 3. Oren, A., Weisburg, W.G., Kessel, M. and Woese, C.R. (1984) Halobacteroides halobius gen. nov., sp. nov., a Moderately Halophic Anaerobic Bacterium from the Bottom Sediments of the Dead Sea. Systematic and Applied Microbiology, 5, 58-70. https://doi.org/10.1016/S0723-2020(84)80051-X
- 4. Suzuki, T., Iwasaki, T., Uzawa, T., Hara, K., Nemoto, N., Kon, T., Ueki, T., Yamagishi, A. and Oshima, T. (2002) Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. Strain 7), a New Member of the Genus Sulfolobus Isolated from Beppu Hot Springs, Japan. Extremophiles, 6, 39-44. https://doi.org/10.1007/s007920100221
- 5. Aguilera, A. (2013) Eukaryotic Organisms in Extreme Acidic Environments, the Río Tinto Case. Life, 3, 363-374. https://doi.org/10.3390/life3030363
- 6. Nguyen, V. A. T., Senoo, K., Mishima, T. and Hisamatsu, M. (2001) Multiple Tolerance of Rhodotorula glutinis R-1 to Acid, Aluminum Ion and Manganese Ion, and Its Unusual Ability of Neutralizing Acidic Medium. Journal of Bioscience and Bioengineering, 92, 366-371. https://doi.org/10.1016/S1389-1723(01)80241-3
- 7. Lu, W., Wen, J., Chen, Y., Sun, B., Jia, X., Liu, M. and Caiyin, Q. (2007) Synergistic Effect of Candida maltosa HY-35 and Enterobacter aerogenes W-23 on Hydrogen Production. Internal Journal of Hydrogen Energy, 32, 1059-1066. https://doi.org/10.1016/j.ijhydene.2006.07.010
- 8. Mitsuya, D., Hayashi, T., Wang, Y., Tanaka, M., Okai, M., Ishida, M. and Urano, N. (2017) Isolation of Aquatic Yeasts with the Ability to Neutralize Acidic Media, from an Extremely Acidic River near Japan’s Kusatsu-Shirane Volcano. Journal of Bioscience and Bioengineering, 124, 43-46. https://doi.org/10.1016/j.jbiosc.2017.02.005
- 9. Okai, M., Suwa, C., Nagaoka, S., Obara, N., Mitsuya, D., Kurihara, A., Ishida, M. and Urano, N. (2017) Neutralization of Acidic Drainage by Cryptococcus sp. T1 Immobilized in Alginate Beads. Bioscience Biotechnology and Biochemistry, 81, 2216-2224. https://doi.org/10.1080/09168451.2017.1373586
- 10. Liu, Y., Tang, H., Lin, Z. and Xu, P. (2015) Mechanisms of acid Tolerance in Bacteria and Prospects in Biotechnology and Bioremediation. Biotechnology Advances, 33, 1484-1492. https://doi.org/10.1016/j.biotechadv.2015.06.001
- 11. Gadanho, M., Libkind, D. and Sampaio, J.P. (2006) Yeast Diversity in the Extreme Acidic Environments of the Iberian Pyrite Belt. Microbial Ecology, 52, 552-563. https://doi.org/10.1007/s00248-006-9027-y
- 12. Nguyen, V. A. T., Tokuda, A., Mishima, T., Nishida, Y. and Hisamatsu, M. (2002) Isolation and Identification of New Acid-Tolerant and Starch-Degradable Yeasts. Journal of Applied Glycoscience, 49, 153-157. https://doi.org/10.5458/jag.49.153
上一篇:Influence of “Mild” Sonicati 下一篇:Antibiotic Resistant Bacteria
最新文章NEWS
- Genomic Recombination Enhances Pathogenic Factors in the Periodontopathogenic Bacterium Eikenella co
- Antibiotic Resistance of Helicobacter pylori and Eradication Rate in Japanese Pediatric Patients
- The PafR Gene Is Required for Antifungal Activity of Strain MS82 against Mycogone perniciosa
- Mycobacteria Interspersed Repetitive Units-Variable Number of Tandem Repeat, Spoligotyping and Drug
- Antimicrobial Activity of Jambul (Syzygium cumini) Fruit Extract on Enteric Pathogenic Bacteria
- Genome Shuffling of <i>Pseudomonas</i> Sp. Ioca11 for Improving Degradation of Polycycli
- Resistance Trends among Pseudomonas aeruginosa Isolates in a Tertiary Care Centre in South Gujarat
- Purification and Characterization of Thermostable Cellulase Free Xylanase from <i>Pseudomonas&
推荐期刊Tui Jian
- Chinese Journal of Integrative Medicine
- Journal of Genetics and Genomics
- Journal of Bionic Engineering
- Pedosphere
- Chinese Journal of Structural Chemistry
- Nuclear Science and Techniques
- 《传媒》
- 《中学生报》教研周刊
热点文章HOT
- Antibiotic Resistance and Potential Pathogenicity of an Isolate Salmonella enterica enterica Based o
- Serotypes, Antibiogram and Genetic Relatedness of Pseudomonas aeruginosa Isolates from Urinary Tract
- Genome Shuffling of <i>Pseudomonas</i> Sp. Ioca11 for Improving Degradation of Polycycli
- Antimicrobial Activity of Jambul (Syzygium cumini) Fruit Extract on Enteric Pathogenic Bacteria
- Resistance Trends among Pseudomonas aeruginosa Isolates in a Tertiary Care Centre in South Gujarat
- Utility of a Relatively Affordable In-House HIV-1 Genotyping Assay for Drug Resistance Testing among
- Antibiotic Resistance of Helicobacter pylori and Eradication Rate in Japanese Pediatric Patients
- Isolation and Characterization of Mercury Resistant Trichoderma Strains from Soil with High Levels o

