Research on the Repeated Sequences among tRNA Sequences
Vol.06No.11(2015), Article ID:61437,7 pages
10.4236/abb.2015.611070
Fangping Wei1*, Yuanze Mao1, Zhenxiong Lan2
1College of Physical Science and Technology, Guangxi University, Nanning, China
2College of Computer and Information Engineering, Guangxi Teacher Education University, Nanning, China

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 11 September 2015; accepted 22 November 2015; published 25 November 2015

ABSTRACT
Many theories thought that present-day tRNA sequences evolved from some short RNA hairpins which contain a simple stem-loop structure. To find out these significant fragment sequences, the repeated sequences of different length within 3420 tRNA sequences are counted and analyzed. The results show that: 1) the probability of occurrence P(i) with the given repeated sequences i follows a power-law distribution when the length K of repeated sequences is longer than four bases, and in this case, the total number N(n) of occurrence with the repeated times n follows a power-law distribution too; 2) the sequence of the length K which repeats the largest times is just only sequence of the length K-b wobbling b bases on its left or right side (b varies between 1 and K-1); 3) the same repeated sequences are found nearly at the identical site in different tRNA sequences as the length K of repeated sequences is longer than five bases. Then a hypothesis of the origin and evolution mechanisms of tRNA sequences is proposed and discussed.
Keywords:
tRNA Sequences, Repeated Sequences, Anticodons

1. Introduction
As we know, the repeated sequence is widespread existed in the genome, which accounts for a large proportion especially in the eukaryotic genomes. Studies have shown that the repeated sequence is of great biological significance. It is important for chromosomes to maintain their space structure, gene expression and gene recombinant [1] [2] . In recent years the studies of repeated sequences turn into a hot issue, many molecular biologists are trying to reveal the structure, function and evolution mechanisms of genes by researching on the repeated sequences. And the repeated sequences have been applied to many fields, e.g. the short tandem repeated sequences are expected to become the second-generation molecular markers.
All modern tRNA sequences evolved from some common ancestral short RNA hairpin [3] -[6] , but their evolutionary mechanism remained an open question. Normally, the gene has the important function must be conservative, then to try to find out the distribution and content of these important fragments of modern tRNA sequences, 3420 tRNA sequences are put as a whole in this paper, and then the repeated sequences of different length within all tRNA sequences are counted. By the analysis of the repeated sequences of all tRNA sequences, the origin and evolution mechanisms of tRNA sequences are discussed further.
2. Materials and Methods
2.1. The Source of tRNA Sequences
The tRNA sequences database was created in 1998 by Sprinzl [7] at first, and it updates constantly day by day. More and more tRNA sequences were collected in this database. All the tRNA sequences used in our paper are downloaded from the database (http://trna.bioinf.uni-leipzig.de/DataOutput/). There are 3719 tRNA sequences in this database altogether which include 61 different anticodons and 429 different species which belonged to three kingdoms respectively: Archaea, Bacteria and Eucarya. Considering the variable loop of the tRNA sequence, then there are 99 bases in each tRNA sequence, and the missing bases are replaced with the line “-” in the tRNA sequence by Sprinzl et al.
2.2. The Method of Counting Repeated Sequences
Firstly, we compare all the 3719 tRNA sequences and remove the high similar or identical sequences, and then just only 3420 tRNA sequences are left and used in our paper. Selecting a fixed length of K string sequences, then various K string sequences of true appearance are counted by us among 3420 tRNA sequences. Considering that there are overlapping within K string sequences and every three bases may represent code information, so we choose three bases as a step when count the K string sequences, such as counting begins with the first base, and once again every three bases until the end of each tRNA sequence. And all the repeated sequences of different length are counted and analyzed.
3. Results and Analysis
3.1. The Repeated Sequences of Different Length with the Highest Occurrences among tRNA Sequences
For the convenience of analysis, just the repeated sequences of the highest occurrences are listed in Table 1. As shown in Table 1, obviously, we can observe that: 1) the highest occurrences of repeated sequences decrease with the increase of the length K. This is because the total number of K string sequences of true appearance within all the tRNA sequences decrease with the increase of the length K. 2) the repeated sequence of the highest occurrences is “TT” which occurs 7183 times when the length K = 2, the repeated sequence “GTT” occurs the most times (3282 times) when K = 3, and the repeated sequence “GTTC” occurs the most times (2080 times) when K = 4. If observe carefully, we find the repeated sequences of the length K with the highest occurrences is just the highest occurrences repeated sequences of the length (K-b) adding b bases on its left or right side (b varies between 1 and (K-1)).This seems to indicate that all the repeated sequences of different length with highest occurrences are at the same site of tRNA sequences and tRNA sequences may put one of the core fragments as a primer to amplify during their formation process. 3) The location of the highest occurrence sequences can be observed in the any arm of tRNA sequence when the length K varies between 1 and 3 bases. However the location of these highest occurrence sequences is nearly observed at the same site when the length K is between 4 and 6 bases, and it fully situates at the same site if only the length K is bigger than 6 bases (see Figure 1). What’s more, the locations of various repeated sequences in the tRNA sequences are counted in this paper. And our results suggest that the same repeated sequences are found nearly at the identical site in the tRNA sequences when the length K of repeated sequences is longer than five bases. 4) The repeated sequences accounts for approximately 82.22% in all the tRNAs. And the longest repeated sequence (AAGATTACCCAAGTCCGGCTG
Table 1. The repeated sequences of different length with highest occurrences. In Table 1, Ac represents acceptor arm, D represents D arm, An represents anticodon arm, E represents extra arm, and T represents T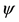 C arm.
C arm.

Figure 1. The location of the most repeated sequence in the secondary structure of tRNA sequences.
AAGGGATCGGTCTTGAAAACCGAGAGTCGG: containing 51 bases) occurs two times, whose anticodon is “TGA” and derived from the Mycoplasma.
In Figure 1, the location of the most repeated sequences is clearly observed in the secondary structure of tRNA sequences. We find that the location of these repeated sequences mainly lies in the anticodon arm and T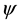 C arm of tRNA sequence. It seems that the repeated sequences take the anticodon arm and T
C arm of tRNA sequence. It seems that the repeated sequences take the anticodon arm and T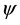 C arm as center and expand towards both directions with the increasing of the length K of repeated sequence (see Figure 1, and the arrow represents the direction of the expansion of repeated sequence). This may indicate that the anticodon arm and T
C arm as center and expand towards both directions with the increasing of the length K of repeated sequence (see Figure 1, and the arrow represents the direction of the expansion of repeated sequence). This may indicate that the anticodon arm and T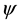 C arm are more significant for tRNA in their evolution process.
C arm are more significant for tRNA in their evolution process.
3.2. The Power-Law Behavior of the Repeated Sequences
The power-law behavior is frequently observed in different fields, such as the population distributions, the social interactions [8] , the World Wide Web [9] and so on. It is also known as Zipf’s law [10] , it was first widely recognized for word usage in text documents. Previous studies [12] -[14] have suggested that the number of distinct parts with a given genomic occurrence followed a power-law distribution. The power-law behavior is observed in our studying of repeated sequences among the 3420 tRNA sequences.
The occurrence frequency of one given repeated sequence i divided by the total number of repeated sequences of true appearance may be taken as the probability of appearance of the repeated sequence i among all the tRNA sequences. As it is shown by Figure 2, the abscissa denotes the repeated sequence i, and the ordinate denotes the probability P(i) of i. Taking into account the paper’s space, therefore we only insert two diagrams into this paper with the length of repeated sequences K = 6 and K = 10. Clearly, Figure 2 shows that the probability P(i) with the given repeated sequence i follows a power-law distribution as the length of repeated sequences K = 6,and K = 10 which means that a few repeated sequences are occurring many times and most occurring infrequently among all the tRNA sequences. What’s more, our researches suggest that the probability P(i) with the given repeated sequence i always follows a power-law distribution when the length of repeated sequences K is longer than four bases.
In Figure 3, the abscissa n denotes the repeated sequences occurring n times, and the ordinate denotes the total number N(n) of repeated sequences which occur n times. As Figure 3 shows, the total number N(n) of repeated sequences which occur n times with the occurrences n follows a power-law distribution too when the length of repeated sequences K = 6 and K = 10. It displays that a few repeated sequences occurring many times and most occurring few times among all the tRNA sequences in these cases as well. Also, the total number N(n) with the occurrences n always follows a power-law distribution when the length of K is longer than four bases.
4. Conclusion and Discussion
The repeated sequences of different length within all the tRNA sequences are counted. Our results show that: 1) the probability P(i) with the given repeated sequences i follows a power-law distribution when the length K of
 (a) (b)
(a) (b)
Figure 2. The probability P(i) of one given repeated sequence i versus the given repeated sequence i. (a) K = 6; (b) K = 10.
 (a) (b)
(a) (b)
Figure 3. The total number N(n) of repeated sequences which occur n times versus the occurrences n. (a) plot of N(n) vs. n when K = 6, (b) plot of P(i) vs. i when K = 8.
repeated sequences is longer than four bases, and in this case, the total number N(n) of repeated sequences which occur n times with the occurrences n follows a power-law distribution too; 2) the highest occurrence sequence of the length K is just only the result of the most repeated sequence of the length K-b wobbling b bases on its left or right side (b varies between 1 and K-1 ); 3) the same repeated sequences are found nearly at the identical site in the different tRNA sequences when the length of repeated sequences is longer than five bases.
Many views have been put on studying the evolutional relationship of tRNA sequences, such as a new tRNA gene may survive through a point mutation in the anticodon sites [15] . Subsequently, the complementary duplication mechanism is also presented as the primary mechanism and point mutation are supporting mechanisms for modern tRNAs’ evolution [16] [17] . So many repeated fragment sequences distribute in the tRNAs, if it hides important information of its evolution? How they arise? We hypothesize that modern tRNA sequences are formed by some fragment sequences acting as primers to duplicate for amplification in their formation process. Supposing that there were only a few fragment sequences in the earliest stage, later the few fragment sequences amplified after replication, and then tRNA sequences didn’t form a stable structure and stopped to amplify until up to their length, or that they could not survive as their length shorter or longer than the length of modern tRNA sequences. Considering the fragment sequences can be affected by the natural environment or suffered AT/GC pressure in the evolution process, the fragment sequences may experience random mutations (such as bases substitution, bases deletion, bases insertion and so on) during evolution, and then the new fragment sequences can be generated [18] [19] . Apart from mutations, Ragan [20] thinks the lateral gene transfer can also be a source of new fragment sequences. Similarly, the new fragment sequences can be used as core primers to duplicate for amplification. And each tRNA sequence must randomly select some fragment sequences as the core primers to duplicate for amplification at first before it turned into a stable molecular structure. In this way, naturally, the higher occurrences of fragment sequences, the more chance of being choose as a core primer to replicate. These fragment sequences underwent selective evolution so a long period that they had resulted in a few repeated sequences occurring many times and most occurring infrequently among all the tRNA sequences and all the tRNA sequences with high similarity in their functions and structures. And the repeated sequences occurring many times may be closer to the earliest fragment sequences.
Our hypothesis of tRNA sequences on the one hand supports the theory that a primitive tRNA consists of seven bases presented by Crick et al. in 1976 [21] and verifies the possibility that tRNA molecule chooses a hairpin RNA as the precursor of tRNA [6] ; on the other hand, our hypothesis not only sustains the view that a hairpin structure is via indirect duplication and then produces another hairpin structure which evolves though base changes, insertions and deletions into the tRNA molecule proposed [5] , but also better supports the model based on a direct duplication of a hairpin structure [22] .
Acknowledgements
This paper was supported by the National Science Foundation of Guangxi of China under Grant Nos: AE052095.
Cite this paper
FangpingWei,YuanzeMao,ZhenxiongLan, (2015) Research on the Repeated Sequences among tRNA Sequences. Advances in Bioscience and Biotechnology,06,669-675. doi: 10.4236/abb.2015.611070
References
- 1. Kang, S. and Janorski, A. (1995) Expansion and Deletion of CTG Repeats from Human Disease Genes Are Determined the Direction of Replication in E. colil. Nature Genetics, 10, 213-218.
http://dx.doi.org/10.1038/ng0695-213 - 2. Jose, A.L. and Lorente, M. (1994) Analysis of Short Tandem Repeat(STR)HUMVWA in the Spanish Population. Forensic Science International, 65, 169-175.
- 3. Bloch, D.P., McArthur, B. and Mirrop, S. (1985) tRNA-rRNA Sequence Homologies: Evidence for an Ancient Modular Format Shared by tRNAs and rRNAs. Biosystems, 17, 209-225.
http://dx.doi.org/10.1016/0303-2647(85)90075-9 - 4. Dick, T.P. and Schamel, W.W.A. (1995) Molecular Evolution of Transfer RNA from Two Precursor Hairpins: Implications for the Origin of Protein Synthesis. Journal of Molecular Evolution, 41, 1-9.
http://dx.doi.org/10.1007/BF00174035 - 5. Eigen, M. and Winkler-Oswatttsch, R. (198l) Transfer-RNA, an Early Gene? Naturwissenschaften, 68, 282-292.
http://dx.doi.org/10.1007/BF01047470 - 6. Hopfield, J.J. (1978) Origin of the Genetic Code: A Testable Hypothesis Based on tRNA Structure, Sequence and Kinetic Proofreading. Proceedings of the National Academy of Sciences USA, 75, 4334-4338.
http://dx.doi.org/10.1073/pnas.75.9.4334 - 7. Sprinzl, M., Horn, C., Brown, M., et al. (1998) Compilation of tRNA Sequence and tRNA Genes. Nucleic Acids Research, 26, 148-153.
http://dx.doi.org/10.1093/nar/26.1.148 - 8. Wasserman, S. and Faust, K. (1994) Social Network Analysis. Cambridge University Press, Cambridge.
http://dx.doi.org/10.1017/CBO9780511815478 - 9. Albert, R., Jeong, H. and Barabasi, A.L. (1999) Internet: Diameter of the World-Wide Web. Nature, 401, 130-131.
http://dx.doi.org/10.1038/43601 - 10. Zipf, G.K. (1949) Human Behaviour and the Principle of Least Effort. Addison-Wesley, Cambridge.
- 11. Qian, J., Luscombe, N.M. and Gerstein, M. (2001) Protein Family and Fold Occurrence in Genomes: Power-Law Behaviour and Evolutionary Model. Journal of Molecular Biology, 313, 673-681.
- 12. Wei F.P. and Lan Z.X. (2007) Using Complex Network to Study the Evolution of tRNA Sequences. Journal of Guangxi University, 32, 246-248.
- 13. Hao, B.-L., Lee, H.C. and Zhang, S.-Y. (2000) Fractals Related to Long DNA Sequences and Complete Genomes. Chaos, Solitons and Fractals, 11, 825-836.
http://dx.doi.org/10.1016/S0960-0779(98)00182-9 - 14. Garg, A., Garg, A. and Tai, K. (2014) A Multi-Gene Genetic Programming Model for Estimating Stress-Dependent Soil Water Retention Curves. Computational Geosciences, 18, 45-56.
http://dx.doi.org/10.1007/s10596-013-9381-z - 15. Saks, M.E., Sampson, J.R. and Abelson, J. (1998) Evolution of a Transfer RNA Gene through a Point Mutation in the Anticodon. Science, 279, 1665-1667.
http://dx.doi.org/10.1126/science.279.5357.1665 - 16. Wei, F.P., Meng, M., Li, S. and Ma, H.R (2006) Comparing Two Evolutionary Mechanisms of Modern tRNAs. Molecular Phylogenetics and Evolution, 38, 1-11.
- 17. Garg, A., Tai, K. and Sreedeep, S. and Stokes, A. (2014) A Computational Intelligence-Based Genetic Programming Approach for the Simulation of Soil Water Retention Curves. Transport in Porous Media, 2014, 1-17.
- 18. Syozo, O. and Jukes, T.H. (1989) Codon Reassignment (Codon Capture) in Evolution. Journal of Molecular Evolution, 28, 271-278.
http://dx.doi.org/10.1007/BF02103422 - 19. Juhling, F. and Morl, M. (2009) tRNAdb 2009: Compilation of tRNA Sequence and tRNA Genes. Nucleic Acids Research, 37, 159-161.
http://dx.doi.org/10.1093/nar/gkn772 - 20. Ragan, M.A. (2001) Detection of Lateral Gene Transfer among Microbial Genomes. Current Opinion in Genetics & Development, 11, 620-626.
http://dx.doi.org/10.1016/S0959-437X(00)00244-6 - 21. Crick, F.H.C., Brenner, S., Klug, A. and Pieczenik, G. (1976) A Speculation on the Origin of Protein Synthesis. Origins Life, 7, 389-397.
http://dx.doi.org/10.1007/BF00927934 - 22. Giulio, M.D. (1992) On the Origin of the Transfer RNA Molecule. Journal of Theoretical Biology, 159, 199-214.
http://dx.doi.org/10.1016/S0022-5193(05)80702-7
NOTES

*Corresponding author.
上一篇:Online Effective Identificatio 下一篇:MicroRNA-21, 204 and 125b Play
最新文章NEWS
- Research on the Repeated Sequences among tRNA Sequences
- MicroRNA-21, 204 and 125b Play Potential Roles in Tumorigenesis of Melanoma
- Agrobacterium-Mediated Transformation of Mexican Lime (Citrus aurantifolia Swingle) Using Optimized
- Quality Control of Selected Antimalarials Sold in the Illicit Market: An Investigation Conducted in
- Structure of Ulvan Isolated from the Edible Green Seaweed, Ulva pertusa
- Effects of Media Composition and Auxins on Adventitious Rooting of Bienertia sinuspersici Cuttings
- Online Effective Identification of Glycopeptide Using Liquid Chromatography Combined with Fourier Tr
- Behavior and Viability of Blueberry Seeds through Germination and Tetrazolium Test
推荐期刊Tui Jian
- Chinese Journal of Integrative Medicine
- Journal of Genetics and Genomics
- Journal of Bionic Engineering
- Pedosphere
- Chinese Journal of Structural Chemistry
- Nuclear Science and Techniques
- 《传媒》
- 《中学生报》教研周刊
热点文章HOT
- Metal Salts Assisted Enzyme-Based Extraction of Stevioside from the Leaves of Stevia rebaudiana Bert
- Efficient Production of δ-Guaiene, an Aroma Sesquiterpene Compound Accumulated in Agarwood, by Meval
- Agrobacterium-Mediated Transformation of Mexican Lime (Citrus aurantifolia Swingle) Using Optimized
- Structure of Alpha-Gliadin Multigene and Construction of Efficient Hairpin RNAi Molecule against Glu
- cDNA Cloning of Paramyosin from Several Kinds of Squid Mantle Muscle
- Biotechnological Transformation of Lignocellulosic Biomass in to Industrial Products: An Overview
- Transforming the Snapdragon Aurone Biosynthetic Genes into Petunia Alters Coloration Patterns in Tra
- Role of Probiotics in Pancreatic Cancer Prevention: The Prospects and Challenges

